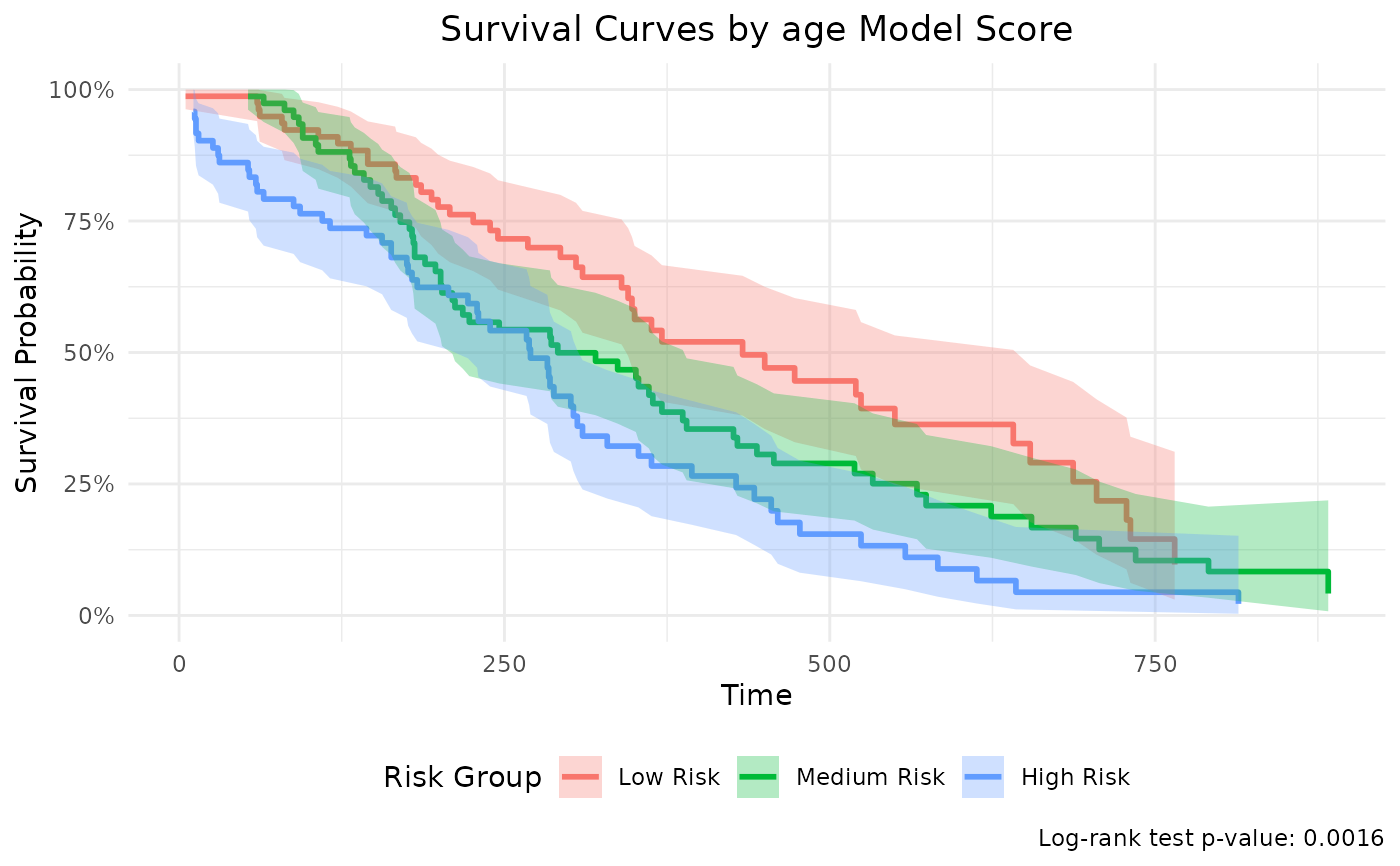
Generate survival curves by grouping observations based on model prediction scores. This function is specifically designed for Cox regression models and creates survival curves comparing different risk groups.
Usage
br_show_survival_curves(
breg,
idx = NULL,
n_groups = 3,
group_labels = NULL,
title = NULL,
subtitle = NULL
)Arguments
- breg
A
bregobject with fitted Cox regression models.- idx
Index or name of the model to use for prediction. If NULL, uses the first model.
- n_groups
Number of groups to create based on score quantiles. Default is 3.
- group_labels
Custom labels for the groups. If NULL, uses "Low", "Medium", "High" for 3 groups or "Q1", "Q2", etc. for other numbers.
- title
Plot title. If NULL, generates automatic title.
- subtitle
Plot subtitle.
Examples
# \donttest{
# Cox regression example with survival curves
if (requireNamespace("survival", quietly = TRUE)) {
lung <- survival::lung |> dplyr::filter(ph.ecog != 3)
mds <- br_pipeline(
lung,
y = c("time", "status"),
x = c("age", "ph.ecog"),
x2 = "sex",
method = "coxph"
)
p <- br_show_survival_curves(mds)
print(p)
}
#> exponentiate estimates of model(s) constructed from coxph method at default
#> `idx` not set, use the first model
 # }
# }
