Arguments
- breg
A regression object with results (must pass
assert_breg_obj_with_results()).- ...
Arguments passing to
br_get_results()for subsetting data table.
See also
Other br_show:
br_show_coxph_diagnostics(),
br_show_fitted_line(),
br_show_fitted_line_2d(),
br_show_forest(),
br_show_forest_circle(),
br_show_forest_ggstats(),
br_show_forest_ggstatsplot(),
br_show_nomogram(),
br_show_residuals(),
br_show_survival_curves(),
br_show_table(),
br_show_table_gt()
Other risk_network:
polar_connect(),
polar_init()
Examples
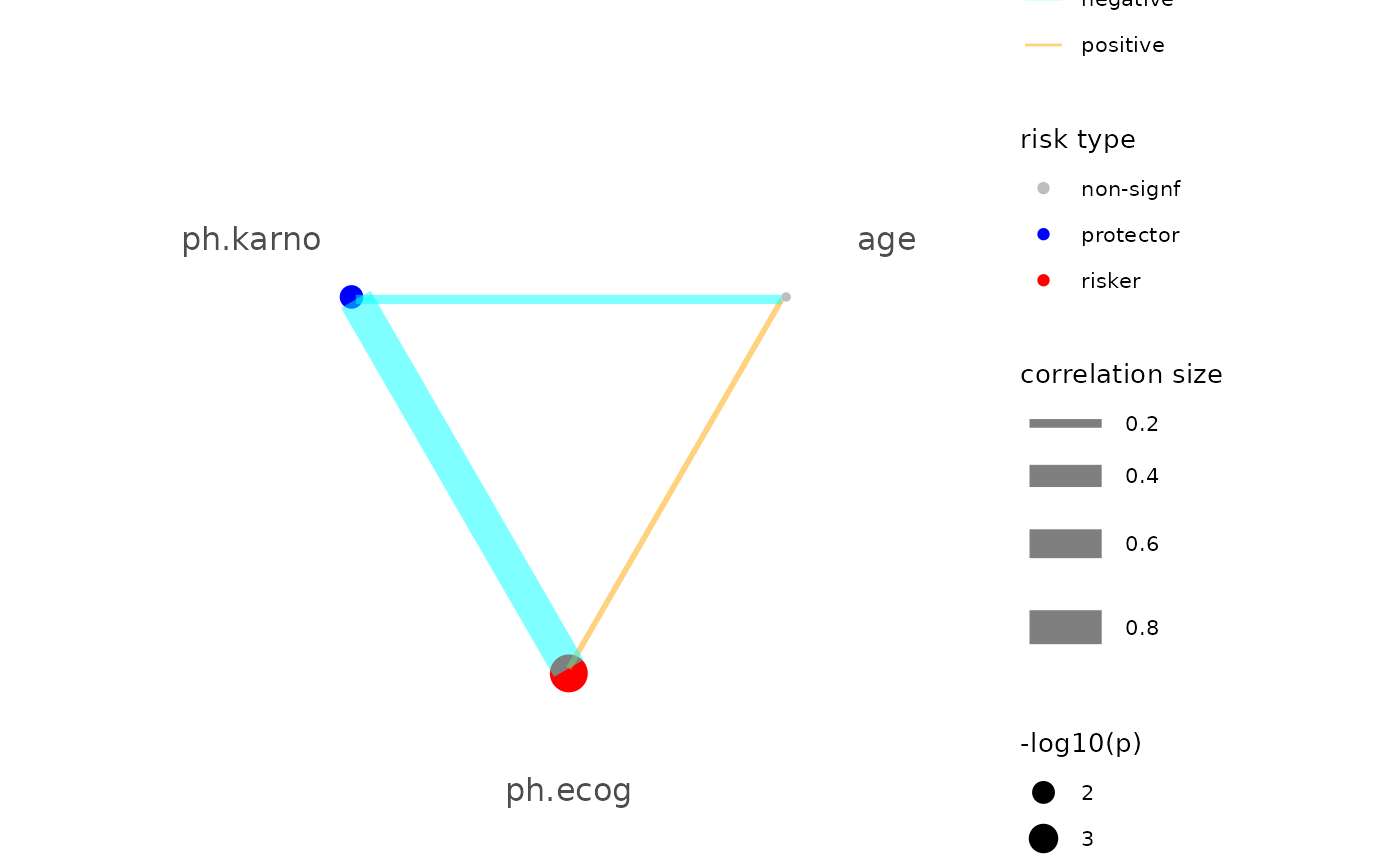
lung <- survival::lung
# Cox-PH regression
mod_surv <- br_pipeline(
data = lung,
y = c("time", "status"),
x = c("age", "ph.ecog", "ph.karno"),
x2 = c("factor(sex)"),
method = "coxph"
)
#> exponentiate estimates of model(s) constructed from coxph method at default
if (requireNamespace("ggnewscale")) {
p <- br_show_risk_network(mod_surv)
p
}
#> Loading required namespace: ggnewscale
#> please note only continuous focal terms analyzed and visualized