This document introduces several auxiliary functions of SigBridgeR.
Add miscellaneous information to the Seurat object
SigBridgeR uses AddMisc() to record what data features
or evidence the various screening algorithms are based on during
execution. As a substitute for the SeuratObject::Misc()
-
AddMisc(): Add miscellaneous information to the Seurat object. Support for adding multiple attributes to theSeuratObject@miscslot simultaneously.
# basic usage
seurat_obj <- AddMisc(seurat_obj, "QC_stats" = qc_df)
# Auto-incrementing example when `cover` set to FALSE
seurat_obj <- AddMisc(seurat_obj, markers = markers1)
seurat_obj <- AddMisc(seurat_obj, markers = markers2, cover=FALSE)
# Add multiple attributes to the `SeuratObject@misc` slot simultaneously
seurat_obj <- AddMisc(seurat_obj, markers1 = markers1, markers2 = markers2)
# Add multiple attributes (stored as a list element) to the `SeuratObject@misc`
seurat_obj <- AddMisc(seurat_obj, list(attr1 = "value1", attr2 = "value2"))Add Gene-level Metadata to the Seurat object
SigBridgeR uses AddMetaFeature() to update the
SeuratObject@assays$RNA@meta.data
Add a new column to the metadata
gene_type <- rep("test", nrow(seurat_obj))
seurat_obj <- AddMetaFeature(seurat_obj, "gene_type" = gene_type)Add a data.frame to the metadata, with the column names as the metadata names.
seurat_obj <- AddMetaFeature(
seurat_obj,
data.frame(gene_type = gene_type, gene_name = rownames(seurat_obj)
)Add to different assays
seurat_obj <- AddMetaFeature(seurat_obj, "gene_type" = gene_type, assay = "RNA")
seurat_obj <- AddMetaFeature(seurat_obj, "gene_type" = gene_type, assay = "ATAC")If duplicate column names are detected, they will be suffixed with an
underscore and a number (e.g., _1, _2) for
disambiguation.
Integration of Single Cell Data
Matrix Integration
When passing raw matrices, the function performs a “join” operation based on the union of all genes. Missing values are filled with NA.
# Create dummy matrices
mat1 <- matrix(rpois(50, 5), nrow = 10, dimnames = list(paste0("G", 1:10), paste0("C", 1:5)))
mat2 <- matrix(rpois(60, 6), nrow = 10, dimnames = list(paste0("G", 5:14), paste0("C", 1:6)))
# Integrate matrices with custom prefixes
integrated_mat <- SCIntegrate(BatchA = mat1, BatchB = mat2)
# default prefixed
integrated_mat2 <- SCIntegrate(mat1, mat2)
integrated_mat[1:6, 1:4]
# BatchA_C1 BatchA_C2 BatchA_C3 BatchA_C4
# G1 3 2 4 5
# G10 2 4 3 4
# G11 NA NA NA NA
# G12 NA NA NA NA
# G13 NA NA NA NA
# G14 NA NA NA NA
integrated_mat2[1:6, 1:4]
# mat1_C1 mat1_C2 mat1_C3 mat1_C4
# G1 3 2 4 5
# G10 2 4 3 4
# G11 NA NA NA NA
# G12 NA NA NA NA
# G13 NA NA NA NA
# G14 NA NA NA NAKey Features:
Duplicate Resolution: Automatically calls
AggregateDupsto handle redundant gene symbols.Auto-Naming: Uses argument names (like BatchA) as cell ID prefixes.
Seurat Integration
For Seurat objects, SCIntegrate automates the standard
workflow via the pipeline parameter.
The Pipeline String: Each letter in the pipeline argument triggers a specific Seurat command:
| Code | Function | Description |
|---|---|---|
| o | CreateSeuratObject |
(do not use it in this function) |
| n | NormalizeData |
Standard normalization. |
| s | ScaleData |
Scales data for PCA. |
| v | FindVariableFeatures |
Selects highly variable genes. |
| p | RunPCA |
Principal Component Analysis. |
| e | FindNeighbors |
Computes SNN graph. |
| c | FindClusters |
Louvain algorithm clustering. |
| t | RunTSNE |
t-SNE reduction. |
| u | RunUMAP |
UMAP reduction. |
| r | SCTransform |
SCT workflow. Replaces n, s, v. |
Example Usage
integrated <- SCIntegrate(
obj1, obj2, # -> merge.Seurat
pipeline = "nsvpi",
method = Seurat::RPCAIntegration, # Change integration method
dims = 1:30, # Passed to RunPCA and IntegrateLayers
k.weight = 50 # Passed to IntegrateLayers
)An example using mock data
mat1 <- matrix(
rpois(1000, 5),
nrow = 20,
dimnames = list(paste0("G", 1:20), paste0("C", 1:50))
)
mat2 <- matrix(
rpois(1200, 6),
nrow = 20,
dimnames = list(paste0("G", 11:30), paste0("C", 1:60))
)
seu1 <- Seurat::CreateSeuratObject(mat1)
seu2 <- Seurat::CreateSeuratObject(mat2)
integrated_seu <- SCIntegrate(
seu1,
seu2,
method = Seurat::CCAIntegration,
pipeline = "nsvpi",
dims = 1:10,
k.weight = 40
)
integrated_seu
# An object of class Seurat
# 30 features across 110 samples within 1 assay
# Active assay: RNA (30 features, 10 variable features)
# 5 layers present: counts.1, counts.2, data.1, data.2, scale.data
# 2 dimensional reductions calculated: pca, integrated.drLoad reference data
Parameters
-
data_type: The type of data to load. Can be either “continuous”, “survival” or “binary”, case-insensitive. -
path: The path to the data directory. -
cache: Whether to cache the data. Defaults toTRUE. -
timeout: The maximum timeout time when downloading data.
When loading the example data, the single-cell RNA expression matrix, the bulk RNA expression matrix, and the clinical phenotype data are loaded all at once. These data are combined into a list and returned.
mydata <- LoadRefData(
data_type = c("survival"),
path = tempdir(),
cache = TRUE,
timeout = 60
)
# * mat_exam (matrix_example)
mydata[[1]][1:6,1:6]
# SMC01.T_AAACCTGCATACGCCG SMC01.T_AAACCTGGTCGCATAT SMC01.T_AAACCTGTCCCTTGCA SMC01.T_AAACGGGAGGGAAACA SMC01.T_AAACGGGGTATAGGTA SMC01.T_AAAGATGAGGCCGAAT
# A1BG 0 0 0 0 0 0
# A1BG.AS1 0 0 0 0 0 0
# A1CF 0 2 0 0 3 0
# A2M 0 0 0 0 0 0
# A2M.AS1 0 0 0 0 0 0
# A2ML1 0 0 0 0 0 0
# * bulk_survival
mydata[[2]][1:6,1:6]
# TCGA-69-7978 TCGA-62-8399 TCGA-78-7539 TCGA-73-4658 TCGA-44-6775 TCGA-44-2655
# HIF3A 4.2598 11.6239 9.1362 5.0288 4.0573 5.5335
# RTN4RL2 8.2023 5.5819 3.5365 7.4156 7.7107 5.3257
# HMGCLL1 2.7476 5.8513 3.8334 3.6447 2.9188 4.8820
# LRRTM1 0.0000 0.4628 4.7506 6.8005 7.7819 2.2882
# GRIN1 6.6074 5.4257 4.9563 7.3510 3.5361 3.3311
# LRRTM3 1.7458 2.0092 0.0000 1.4468 0.0000 0.0000
# * pheno_survival
mydata[[3]] |> head()
# time status
# TCGA-69-7978 4.40 0
# TCGA-62-8399 88.57 0
# TCGA-78-7539 25.99 0
# TCGA-73-4658 52.56 1
# TCGA-44-6775 23.16 0
# TCGA-44-2655 43.50 0We recommend using the zeallot package’s
%<-% function to assign values and rename them
simultaneously.
Setting up Python Environment
Some screening methods (e.g. Section 3.5 DEGAS) are built using Python and require an execution environment. Here is a function to help you set up a Python environment. Both Windows and Unix-like systems are supported.
# * This is an example of setting up a Python environment using conda
SetupPyEnv(
env_type = "conda",
env_name = "test-condaenv",
method = c("reticulate", "system","environment"), # choose one of them, default is "reticulate"
env_file = NULL # path to environment.yml file, used when method = "environment"
python_version = NULL,
packages = c(
"pandas" = "1.3",
"numpy" = "any"
),
recreate = FALSE, # whether to remove the existing environment and recreate it
use_conda_forge = TRUE,
verbose = TRUE
)
reticualte::use_condaenv("test-condaenv")
# * Or use virtualenv via reticulate
SetupPyEnv(
env_type = "venv",
env_name = "test-venv",
python_version = "3.9.15",
packages = c("tensorflow" = "2.4.1", "protobuf" = "3.20.3"),
python_path = NULL,
recreate = FALSE,
verbose = TRUE
)
reticulate::use_virtualenv("test-venv")You can use ListPyEnv() to list all the Python
environments you have set up. Both conda and virtual environments are
supported.
# * Unix-like systems
ListPyEnv()
# name python type
# 1 base /home/user/miniconda3/bin/python conda
# 2 test-condaenv /home/user/miniconda3/envs/test-condaenv/bin/python conda
# 3 test-venv /home/user/miniconda3/envs/test-venv/bin/python venvShow the conda environments only:
# * Unix-like systems
ListPyEnv(env_type = "conda")
# name python type
# 1 base /home/user/miniconda3/bin/python conda
# 2 test-condaenv /home/user/miniconda3/envs/test-condaenv/bin/python condaIf the virtual environment isn’t installed in the default location,
you can specify the location of the virtual environment with the
venv_locations parameter.
ListPyEnv(env_type = "venv", venv_locations ="~/here_is_a_dir/.virtualenvs")Finding the Optimal Number of Principle Components
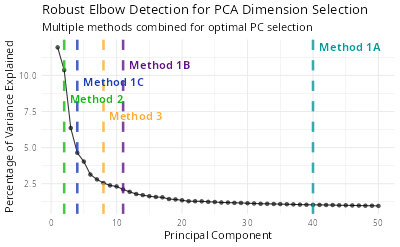
Usually the number if principle components (PCs) is manually set to 10 or 20 according to the elbow plot. However, it is not always the case that the PCs with the highest variance are the most informative. Here is a function to help you find the optimal number of PCs.
ndims <- FindRobustElbow(
obj = seurat,
verbose = TRUE,
ndims = 50
)
# ── Method Results
# ℹ Method 1A (Cumulative Variance > 90%): 1:40
# ℹ Method 1B (Variance > Mean): 1:11
# ℹ Method 1C (Variance > 2*SD): 1:4
# ℹ Method 2 (Second Derivative): 1:2
# ℹ Method 3 (Distance-based): 1:8
# ✔ Final Recommended Dimensions: 1:35
# ── Summary
# ℹ Cumulative variance at 35 PCs: 84.8%
# ℹ Variance explained by PC35: 1.09%
print(ndims)
# 35This function will draw an elbow plot with each method and the
recommended number of PCs. You can also use the ndims
parameter to specify the maximum number of PCs to be tested. The default
value is 50.
knitr::include_graphics("vignettes/example_figures/elbow.png")Setting and Retriving Package Options
Currently, 7 package options are provided:
-
verbose: Whether to print the progress of the function. Default isTRUE. -
seed: The random seed used in the function. Default is123L. -
timeout: The maximum timeout time when downloading data. Default is180L.
These options can be modified using either the
setFuncOption or Options function (the former
is recommended).
setFuncOption(seed = 321L)
options(SigBridgeR.seed = 321L)setFuncOption automatically detects the prefix, so
parameters with a package name prefix are also compatible.
setFuncOption(SigBridgeR.seed = 321L)getFuncOption is used to retrieve the global parameters
of SigBridgeR.
getFuncOption("seed")
# 321
# * Auto-detect prefix
getFuncOption("SigBridgeR.seed")
# 321