Full Tutorial for SigBridgeR
0. Preface
0.1 Contents
0.1 Introduction to SigBridgeR
SigBridgeR (short for Significant cell-to-phenotype Bridge in R) is an R package for screening cells highly associated with phenotype data using single-cell RNA-seq, bulk RNA expression and sample related phenotype data (e.g. patient survival, age, etc). It integrates many single cell phenotypic screening methods (8. References) and provides unified preprocessing, parameter tuning, and visualization approaches,performing as a unified integration panel.
1. Installation
Install SigBridgeR using one of these methods:
1.1 Release from GitHub
Usually we recommend installing the latest release from GitHub because of the latest features and bug fixes.
if(!requireNamespace("pak")) {
install.packages("pak")
}
pak::pkg_install("WangLabCSU/SigBridgeR")1.2 Release from r-universe
install.packages("SigBridgeR", repos = "https://wanglab.r-universe.dev")If you encounter compatibility issues, you can install the version of the package indicated in 7. Troubleshooting.
2. Loading and preprocessing data
First load the packages, you will see a version number message indicating successful loading:
library(SigBridgeR)
# ✔ SigBridgeR v2.5.2 loaded2.1 Single-cell RNA-seq Data
You can use function SCPreProcess to preprocess your
single-cell RNA-seq data. Here are some options:
2.1.1 (Option A) Start from Raw Matrix
When starting from a raw count matrix (data.frame, matrix or
dgCMatrix), SCPreProcess will automatically perform Seurat
preprocess (including
NormalizeData, FindVariableFeatures, ScaleData, RunPCA, FindNeighbors, FindClusters, RunTSNE, RunUMAP).
SCPreProcess returns a fully preprocessed Seurat object for
downstream use.
your_seurat <- SCPreProcess(
your_matrix,
# Parameters used in Seurat preprocessing pipeline
meta_data = NULL,
project = "Screen_Single_Cell",
min_cells = 400,
min_features = 0,
normalization_method = "LogNormalize",
scale_factor = 10000,
scale_features = NULL,
selection_method = "vst",
resolution = 0.6,
dims = 1:10,
verbose = TRUE,
# Parameters used in SigBridgeR workflow
future_global_maxsize = 6 * 1024^3,
quality_control = TRUE,
quality_control.pattern = c("^MT-"), # human for '^MT-', mouse for '^mt-', or specify your own pattern
data_filter = TRUE,
data_filter.nFeature_RNA_thresh = c(200, 6000), # [min, max]
data_filter.percent.mt = 20,
column2only_tumor = NULL,
...
)-
Build the initial Seurat object
- Imports the count matrix.
- Applies the first, coarse filters: genes must be detected in >=
min_cellscells; cells must contain >=min_featuresgenes.
- Attaches optional sample metadata.
- Imports the count matrix.
-
Mitochondrial QC (default: ON)
- A single regular expression (
^MT-or^mt-) is used to compute the percentage of mitochondrial reads per cell. Customized regexps are also supported. - The metric is stored in
object$percent.mtfor later filtering.
- A single regular expression (
-
Cell filtering (default: ON)
Cells are retained only if they satisfy all of the following:-
nFeature_RNAlies between the lower and upper bounds defined bydata_filter.nFeature_RNA_thresh.
-
percent.mtis below the user-defined cut-off (data_filter.percent.mt).
-
-
Normalisation, variable-feature selection and scaling
Encapsulated inProcessSeuratObject():- Counts are normalised with the chosen method (default:
LogNormalize).
- Highly variable features are identified using the requested
algorithm (default: vst).
- Expression values of those features are scaled to unit variance.
- Counts are normalised with the chosen method (default:
LogNormalize).
-
Dimensionality reduction and clustering
ClusterAndReduce()runs:- PCA -> nearest-neighbour graph -> Louvain clustering
(resolution parameterised).
- UMAP for two-dimensional visualization.
- Optionally removes very small or low-quality clusters.
- PCA -> nearest-neighbour graph -> Louvain clustering
(resolution parameterised).
Tumour-cell flagging
FilterTumorCell()use a binary label (Tumour, Normal) from the column specified bycolumn2only_tumorand filters the Seurat object to contain only the cancer cells.
2.1.2 (Option B) Start from AnnDataR6 Object
SCPreProcess also supports AnnData objects. You may
reference and use the following code:
reticulate::use_pythonenv("The_path_to_your_python")
# reticulate::use_condaenv("conda_env_name")
anndata_obj <- anndata::read_h5ad("path_to_your_file.h5ad") # Or other file formats, make sure the matrix is in obj$X.
your_seurat <- SCPreProcess(
anndata_obj,
# Parameters used in Seurat preprocessing pipeline
meta_data = NULL,
project = "Screen_Single_Cell",
min_cells = 400,
min_features = 0,
normalization_method = "LogNormalize",
scale_factor = 10000,
scale_features = NULL,
selection_method = "vst",
resolution = 0.6,
dims = 1:10,
verbose = TRUE,
# Parameters used in SigBridgeR workflow
future_global_maxsize = 6 * 1024^3,
quality_control = TRUE,
quality_control.pattern = c("^MT-"), # human for '^MT-', mouse for '^mt-', or specify your own pattern
data_filter = TRUE,
data_filter.nFeature_RNA_thresh = c(200, 6000), # [min, max]
data_filter.percent.mt = 20,
column2only_tumor = NULL,
...
)The description of data (meta.data) in anndata_obj$obs
will be add to your_seurat@meta.data.
helpful documentation:
2.1.8 (Optional) Filter Out Tumor Cells
If you aim to filter out phenotype-associated cells in all tumor
cells. With a preprocessed Seurat object (containing results from ),
SCPreProcess will filter out tumor cells using the
specified metadata column:
your_seurat <- SCPreProcess(your_seurat, column2only_tumor = "Tissue")Note: I don’t recommend using columns like
column2only_tumor = "Celltype"as tumor cell identities vary across tissues. instead:
Create a Dedicated Column: Add a new metadata column (e.g., is_tumor) to explicitly label cells:“Tumo(u)r”/“Normal”
Code Example:
2.2 Bulk expression data
Here are some methods for processing bulk RNA-seq gene expression data matrices.
2.2.1 Evaluate the quality of your bulk RNA-seq data
BulkPreProcess performs comprehensive quality control on
bulk RNA-seq data.
Key parameters for BulkPreProcess:
-
data: Expression matrix with genes as rows and samples as columns, or a list containing count_matrix and sample_info. -
sample_info: Sample information data frame (optional), ignored if data is a list. A qualifiedsample_infoshould contain bothsampleandconditioncolumns (case-sensitive), and there are no specific requirements for the data type stored in theconditioncolumn. -
gene_symbol_conversion: Whether to convert Ensembl version IDs and TCGA version IDs to genes with SymbolConvert in Section 2.2.2, default TRUE. -
check: Whether to perform detailed quality checks, default TRUE. -
min_count_threshold: Minimum count threshold for gene filtering, default 10. -
min_gene_expressed: Minimum number of samples a gene must be expressed in, default 3. -
min_total_reads: Minimum total reads per sample, default 1e6. -
min_genes_detected: Minimum number of genes detected per sample, default 10000. -
min_correlation: Minimum correlation threshold between samples, default 0.8. -
n_top_genes: Number of top variable genes for PCA analysis, default 500. -
show_plot_results: Whether to generate PCA visualization plots, default TRUE. -
verbose: Whether to output detailed information, default TRUE.
Suppose you have bulk RNA-seq count data and sample information, and you want to perform comprehensive preprocessing and quality control. You can refer to and use the following code:
# Example usage of BulkPreProcess
filtered_counts <- BulkPreProcess(
data = your_count_matrix,
sample_info = your_sample_info,
gene_symbol_conversion = TRUE,
check = TRUE,
min_count_threshold = 10,
min_samples_expressed = 3,
min_total_reads = 1e6,
min_genes_detected = 10000,
min_correlation = 0.8,
n_top_genes = 500,
show_plot_results = TRUE,
verbose = TRUE
)The function returns a filtered bulk count matrix after applying quality control steps.
Quality Control Metrics Reported
-
Gene Filtering:
First, lowly expressed genes are removed, retaining genes that have an expression level reaching
min_count_thresholdin at leastmin_gene_expressedsamples. This step is based on statistical power considerations, as lowly expressed genes are prone to false-positive results in differential analysis. -
Sample Filtering:
Subsequently, low-quality samples are filtered based on three core indicators:
- Sequencing Depth: The total number of reads must reach
min_total_reads. - Library Complexity: The number of detected genes must exceed
min_genes_detected. - Sample Consistency: When quality checking is enabled, the average
correlation between samples must meet the
min_correlationthreshold.
- Sequencing Depth: The total number of reads must reach
Data Integrity Check: Identifies technical anomalies; too many missing values may indicate experimental issues.
Feature Expression Filtering: Uses a count threshold rather than a proportion threshold to avoid over-penalizing small samples. Genes must be stably expressed in multiple samples to ensure the reliability of statistical tests.
-
Sample-Level Filtering:
Comprehensively evaluates technical quality:
Sequencing depth ensures detection sensitivity, the number of detected genes reflects library diversity, and sample correlation (Pearson) validates experimental reproducibility. PCA analysis further identifies batch effects and outlier samples. -
PCA Variance:
Variance explained by first two principal components.
Whenshow_plot_results = TRUE, the function generates PCA plot colored by experimental condition Batch Effects: Proportion of genes significantly affected by batch
Recommended Parameter Adjustments
Parameter settings need to balance data quality and information retention:
-
For Low-Depth Sequencing Data:
Should relax
min_total_readsandmin_genes_detecteddue to technical limitations rather than quality issues causing low indicators. -
For Noisy Datasets:
Need to increase the
min_correlationthreshold to strengthen sample consistency requirements and exclude technical variation interference. -
For Large Datasets:
Consider setting
check = FALSEto enable only partial filtering functions and speed up the process.
All parameters are set based on empirical values, and users should make appropriate adjustments according to the specific experimental design and sequencing platform characteristics.
2.2.2 Gene Symbol Conversion
SymbolConvert performs a straightforward task:
converting common gene identifiers (e.g., Ensemble IDs, Entrez) to
standardized gene symbols by using the IDConverter
package.
# genes * samples
your_bulk_data <- read.csv("path_to_your_file.csv", header = TRUE, row.names = 1)
your_bulk_data <- SymbolConvert(your_bulk_data)You can also use other package like org.Hs.eg.db for
gene symbol matching if you prefer not to use SymbolConvert’s built-in
IDConverter.
library(org.Hs.eg.db)
your_bulk_data <- read.csv("path_to_your_file.csv", header = TRUE, row.names = 1)
your_bulk_data <- BulkPreProcess(your_bulk_data, gene_symbol_conversion = FALSE)
ensembl_ids <- sub("\\..*", "", rownames(your_bulk_data))
gene_symbols <- mapIds(org.Hs.eg.db,
keys = ensembl_ids,
column = "SYMBOL",
keytype = "ENSEMBL",
multiVals = "first")
rownames(your_bulk_data) <- gene_symbols2.3 Phenotype Data
Bascially you can just use your phenotype data directly. If you are
confused about the structure Screen() requires, please
refer to Section 5.
You can use this function to check NA values, which tells you the location of NA values in your phenotype data:
# `max_print`: output how many NA location messages at one time if NA exists
CheckNA <- function(data, max_print = 5) {
# Load required packages
rlang::check_installed(c("dplyr", "purrr", "cli", "data.table"))
# Convert to data.table if it's a 2D structure but not already a data.table
is_2d <- !is.null(dim(data))
dt <- NULL
if (is_2d && !data.table::is.data.table(data)) {
dt <- data.table::as.data.table(data)
} else if (is_2d) {
dt <- data
}
# Initialize output
na_info <- list()
cli::cli_h1("NA Value Check (data.table optimized)")
cli::cli_alert_info("Checking object with dimensions: {.val {dim(data)}}")
# Handle 1D data (vectors)
if (!is_2d) {
na_count <- sum(is.na(data))
na_positions <- which(is.na(data))
na_info$positions <- na_positions
na_info$count <- na_count
if (na_count == 0) {
cli::cli_alert_success("No NA values found in the vector!")
}
cli::cli_alert_warning("Found {.val {na_count}} NA value{?s} in vector")
if (length(na_positions) > 0 && max_print > 0) {
positions_to_show <- na_positions[
1:min(max_print, length(na_positions))
]
cli::cli_text("Positions: {.val {positions_to_show}}")
if (length(na_positions) > max_print) {
cli::cli_text(
"{.val {length(na_positions) - max_print}} additional positions not shown"
)
}
}
# For named vectors
if (!is.null(names(data))) {
na_names <- names(data)[na_positions]
na_info$names <- na_names
if (length(na_names) > 0 && max_print > 0) {
names_to_show <- na_names[1:min(max_print, length(na_names))]
cli::cli_text("Names: {.val {names_to_show}}")
}
}
} else {
# Handle 2D data (dataframes, matrices, data.tables)
# Use data.table for efficient NA counting
na_count <- dt[, sum(is.na(.SD))]
na_info$count <- na_count
if (na_count == 0) {
cli::cli_alert_success("No NA values found in the 2D data!")
}
cli::cli_alert_warning(
"Found {.val {na_count}} NA value{?s} in 2D data"
)
# Get NA positions efficiently without melt warning
if (max_print > 0) {
# Create a matrix of logical values indicating NA positions
na_matrix <- is.na(as.matrix(dt))
na_positions <- which(na_matrix, arr.ind = TRUE)
# Convert to data.table and add column names
na_positions_dt <- data.table::as.data.table(na_positions)
colnames(na_positions_dt) <- c("row", "col")
na_positions_dt$col_name <- colnames(dt)[na_positions_dt$col]
na_info$positions <- na_positions_dt
if (nrow(na_positions_dt) > 0) {
positions_to_show <- na_positions_dt[
1:min(max_print, nrow(na_positions_dt)),
]
cli::cli_text(
"First {.val {nrow(positions_to_show)}} position{?s}:"
)
for (i in 1:nrow(positions_to_show)) {
cli::cli_text(
" Row {.val {positions_to_show$row[i]}}, Col {.val {positions_to_show$col_name[i]}} (index: {.val {positions_to_show$col[i]}})"
)
}
if (na_count > max_print) {
cli::cli_text(
"{.val {na_count - max_print}} additional positions not shown"
)
}
}
}
# Column-wise summary using data.table
col_na <- dt[, lapply(.SD, function(x) sum(is.na(x)))]
col_na <- unlist(col_na)
col_na <- col_na[col_na > 0]
if (length(col_na) > 0) {
na_info$column_na <- col_na
cli::cli_text("Column-wise NA counts:")
for (col in names(col_na)) {
cli::cli_text(" {.field {col}}: {.val {col_na[col]}}")
}
}
}
return(invisible(na_info))
}3. Screening Cells Associated with Phenotype
The function Screen provide 5 different
options for screening cells associated with phenotype, These 5
algorithms come from the repositories mentioned in Section 0.1, and you can
choose one of them to screen your cells.
Key parameters for Screen:
-
matched_bulk: A data frame of bulk expression data after intersecting samples. -
sc_data: A Seurat object after preprocessing, you can use the output ofPreprocessfunction or your own preprocessed Seurat object. -
phenotype: A data frame of phenotype data after intersecting samples. -
label_type: A character value specifying the filtering labels are stored in theSeurat_object@misc, default:NULL. -
phenotype_class: A character value specifying the phenotype data type, i.e."binary","survival"or"continuous". -
screen_method: A character value specifying the screening method, i.e. “Scissor”, “scPAS”, “scAB” or “scPP” -
...: Other parameters for the screening methods.
3.1 (Option A) Scissor Screening
Parameters pass to ... when using Scissor
method:
-
path2save_scissor_inputs: A character value specifying the path to save intermediate data, you can also setpath2load_scissor_cache=NULLto suppress the saving of intermediate files. Default:Scissor_inputs.RData -
path2load_scissor_cahce: A character value specifying the path to load intermediate data -
alpha: Parameter used to balance the effect of the l1 norm and the network-based penalties. It can be a number or a searching vector. If alpha = NULL, a default searching vector is used. The range of alpha is in[0,1]. A larger alpha lays more emphasis on the l1 norm. -
cutoff: Cutoff for the percentage of the Scissor selected cells in total cells. This parameter is used to restrict the number of the Scissor selected cells. A cutoff less than 50% (default 20%) is recommended depending on the input data. -
reliability_test: A logical value specifying whether to perform reliability test. Default:FALSE -
reliability_test.n: Permutation times (default: 10) -
reliability_test.nfold: The fold number in cross-validation (default: 10) -
cell_evaluation: A logical value specifying whether to perform cell evaluation. Default:FALSE -
cell_evaluation.benchmark_data: Path to benchmark data (RData file). -
cell_evaluation.FDR: FDR threshold for cell evaluation (default: 0.05). -
cell_evaluation.bootstrap_n: Number of bootstrap iterations for cell evaluation (default: 100).
Usage:
scissor_result = Screen(
matched_bulk = matched_bulk,
sc_data = sc_dataset, # A Seurat object after preprocessing
phenotype = matched_phenotype_data,
label_type = "TP53", # The filtering labels are stored in the `@misc`, you can change it to your own label
phenotype_class = "binary",
screen_method = c("Scissor"),
path2save_scissor_inputs = "Tmp/Scissor_inputs.RData" # Intermediate data
)You can use the intermediate data for repeated runs. This is an
inherent feature of the scissor.
scissor_result = Screen(
sc_data = sc_dataset,
label_type = "TP53",
phenotype_class = "binary",
screen_method = c("Scissor"),
path2load_scissor_cahe = "Tmp/Scissor_inputs.RData" # Intermediate data
)If only the parameters alpha and cutoff are
adjusted, this method can also be applied.
# When `alpha = NULL`, an alpha iteration will continue until phenotype-associated cells are screened out or no cells are screened out even after exceeding the `cutoff`.
scissor_result = Screen(
sc_data = sc_dataset,
label_type = "TP53",
phenotype_class = "binary",
screen_method = c("Scissor"),
path2load_scissor_cahce = "Tmp/Scissor_inputs.RData", # Intermediate data
alpha = NULL,
cutoff = 0.05
)Returning structure: A list containing:
-
scRNA_data: A Seurat object after screening -
scissor_result: The result of Scissor screening, including the parameters used -
reliability_test: Reliability test results -
cell_evaluation: Cell evaluation results
Cell level Evaluation & Reliability Test:
You can use cell_evalutaion = TRUE and
reliability_test = TRUE to obtain some supporting
information for each Scissor selected cell. First, prepare a benchmark
dataset yourself for cell evalutaion.
scissor_result = Screen(
sc_data = sc_dataset,
label_type = "TP53",
phenotype_class = "binary",
screen_method = c("Scissor"),
path2load_scissor_cahce = "Tmp/Scissor_inputs.RData", # Intermediate data
reliability_test = TRUE,
cell_evaluation = TRUE,
cell_evaluation.benchmark_data = "path_to_benchmark_data.RData",
alpha = NULL,
cutoff = 0.05
)helpful documentation:
3.2 (Option B) scPAS Screening
Parameters pass to ... when using scPAS
method (basically adapted from the scPAS’s
documentation):
-
Parameters passed to
scPAS::scPAS()These parameters directly interface with the core
scPAS() function from the original package:-
assay: Name of Assay to get. -
imputation: Logical. imputation or not. -
nfeature: Numeric. The Number of features to select as top variable features insc_data. Top variable features will be used to intersect with the features ofmatched_bulk. Default is NULL and all features will be used. -
alpha: Numeric. Parameter used to balance the effect of the l1 norm and the network-based penalties. It can be a number or a searching vector. Ifalpha = NULL, a default searching vector is used. The range of alpha is in[0,1]. A larger alpha lays more emphasis on the l1 norm. -
cutoff: Numeric. Cutoff for the percentage of the scPAS selected cells in total cells whenalpha = NULL. This parameter is used to restrict the number of the scPAS selected cells. A cutoff less than 50% (default 20%) is recommended depending on the input data. -
network_class: The source of feature-feature similarity network. By default this is set to sc and the other one is bulk.
-
usage:
scpas_result = Screen(
matched_bulk = matched_bulk,
sc_data = A_Seurat_object,
phenotype = phenotype,
label_type = "TP53", # The filtering labels are stored in the `@misc`
screen_method = "scpas",
phenotype_class = "binary"
)returning structure: A list containing:
-
scRNA_data: A Seurat object after screening -
stats: A data.frame the significance of scPAS screening results -
para: A list containing the parameters used in scPAS screening
3.3 (Option C) scAB Screening
Parameters pass to ... when using scAB
method (basically adapted from the scAB’s
documentation):
-
alpha: Coefficient of phenotype regularization, default is0.005 -
alpha_2: Coefficient of cell-cell similarity regularization, default is5e-05 -
maxiter: Maximum number of iterations, default is2000 -
tred: Threshold for early stopping, default is2
usage:
scab_result = Screen(
matched_bulk = your_matched_bulk,
sc_data = A_Seurat_object,
phenotype = your_matched_phenotype,
label_type = "TP53", # The filtering labels are stored in the `@misc`
screen_method = "scAB",
phenotype_class = "binary",
)returning structure: A list containing:
-
scRNA_data: A Seurat object after screening -
scAB_result: A list with the submatrix and loss value
3.4 (Option D) scPP Screening
Parameters pass to ... when using scPP
method :
-
ref_group: The reference group for the binary analysis, default is1 -
Log2FC_cutoff: The cutoff for the log2 fold change of the binary analysis, default is0.585 -
estimate_cutoff: Effect size threshold for continuous traits, default is0.2 -
probs: Quantile cutoff for cell classification, default is0.2
usage:
# This will take several hours
scpp_result = Screen(
matched_bulk = your_matched_bulk,
sc_data = A_Seurat_object,
phenotype = your_matched_phenotype,
label_type = "TP53", # The filtering labels are stored in the `@misc`
screen_method = "scpp",
phenotype_class = "binary",
)returning structure: A list containing:
-
scRNA_data: A Seurat object after screening -
gene_list: A list containing positive genes and negative genes -
AUC: A data.frame with area under the ROC curve
3.5 (Option E) DEGAS Screening
Parameters pass to ... when using DEGAS
method
-
sc_data.pheno_colname: The column name of the phenotype in thesc_data@meta.dataslot, used to specify the phenotype for more accurate screening. Default isNULL. -
tmp_dir: The directory for storing the intermediate files, default isNULL. -
env_params: A list of parameters for the environment, default islist(). Use?DoDEGASto see the details. -
degas_params: A list of parameters for the DEGAS algorithm, default islist(). Use?DoDEGASto see the details. -
normality_test_method: Method for normality testing:"jarque-bera", "d'agostino", or "kolmogorov-smirnov", default is “jarque-bera”.
degas_result = Screen(
matched_bulk = your_matched_bulk,
sc_data = A_Seurat_object,
phenotype = your_matched_phenotype,
label_type = "TP53", # The labels are stored in the `@misc` and are used to identify the screening results.
screen_method = "DEGAS",
# The type of phenotype
phenotype_class = c("binary", "continuous", "survival"),
# Environment parameters
env_params = list(
env.name = "r-reticulate-degas",
env.type = "conda",
# Environment.yml file will be used to create the conda environment in default, so other parameters can be omitted
env.method = "environment",
# The path of the environment.yml file
env.file = system.file(
"conda/DEGAS_environment.yml",
package = "SigBridgeR"
),
env.python_verion = "3.9.15",
env.packages = c(
"tensorflow" = "2.4.1",
"protobuf" = "3.20.3"
),
env.recreate = FALSE,
env.use_conda_forge = TRUE,
env.verbose = FALSE
),
# DEGAS parameters
degas_params = list(
# DEGAS.model_type will be automatically determined by the `phenotype_class`
DEGAS.model_type = c(
"BlankClass", # only bulk level phenotype specified
"ClassBlank", # only single cell level phenotype specified
"ClassClass", # when both single cell level phenotype and bulk level phenotype specified
"ClassCox", # when both single cell level phenotype and bulk level survival data specified
"BlankCox" # only bulk level survival data specified
),
# DEGAS.architecture will be `DenseNet` in default
DEGAS.architecture = c(
"DenseNet", # a dense net network
"Standard" # a feed forward network
),
# The path to save intermediate data
path.data = '',
path.result = '',
# The python executable path of the conda environment, auto detected in default
DEGAS.pyloc = NULL,
# Some functions will be called in the DEGAS algorithm
DEGAS.toolsPath = paste0(.libPaths()[1], "/DEGAS/tools/"), # or `file.path(.libaPaths()[1], "SigBridgeR/DEGAS_tools/")`
# Screening parameters
DEGAS.ff_depth = 3,
DEGAS.bag_depth = 5,
DEGAS.train_steps = 2000,
DEGAS.scbatch_sz = 200,
DEGAS.patbatch_sz = 50,
DEGAS.hidden_feats = 50,
DEGAS.do_prc = 0.5,
DEGAS.lambda1 = 3.0,
DEGAS.lambda2 = 3.0,
DEGAS.lambda3 = 3.0,
DEGAS.seed = 2
),
# default: "jarque-bera".
normality_test_method = c(
"jarque-bera",
"d'agostino",
"kolmogorov-smirnov"
)
)This code chunk will CREATE a conda environment called
“r-reticulate-degas” with python 3.9.15, and install
the required packages (tensorflow, protobuf) for DEGAS. If an
environment with the same name already exists, it will directly use this
environment without creating a new one (unless specified
env_params = list(env.recreate=TRUE)).
You can use ListPyEnvs() to list all the python
environments in your system, including virtual environments. Both
Windows and Unix-like systems are supported. More information can be
found in Section
6.3
# * On Unix-like system it goes like this
ListPyEnv()
# name python type
# 1 base /home/user/miniconda3/bin/python conda
# 2 r-reticulate-degas /home/user/miniconda3/envs/r-reticulate-degas/bin/python conda
# 3 test /home/user/.virtualenvs/test/bin/python venvPlease note that the environmental dependencies required for DEGAS to run are quite stringent, and conflicts are highly likely to occur. This is why the use of this screening method is not recommended.
returning structure: A list containing:
-
scRNA_data: A Seurat object after screening -
model: A model trained using single-cell RNA expression matrix, tissue bulk RNA sequencing expression matrix, and phenotypic data. -
DEGAS_prediction: Using the model to conduct prediction for each cell, resulting in a data.frame where each phenotype has a predicted probability score.
3.8 Merge screening results
If you have performed multiple screening methods one the same
single-cell data, you can use the MergeResult to merge the
screening results of these methods. The Seurat object or a results list
from Screen is accepted.
merged_seurat = MergeResult(
your_scissor_result,
your_scPAS_result,
your_scAB_result,
your_scPP_result,
your_DEGAS_result
)
# * mixed input form is alse supported
merged_seurat = MergeResult(
your_scissor_result$scRNA_data,
your_scPAS_result$scRNA_data,
your_scAB_result,
your_scPP_result,
your_DEGAS_result
)This function simply merges the meta.data and
misc slots of the input Seurat objects. Please note that
the intermediate data (e.g.,
scissor_result$reliability.test or
scab_result$scAB_result) will not be preserved in this
process.
returning structure:
A Seurat object with merged meta.data and
misc slots.
4. Visualization
Here we provide some visualization methods for the screening results.
Considering that many people have different needs for data
visualization, SigBridgR hardly provides visualization
(except for fraction plot and upset plot, because we provide some
statistic results for them). We only provide the source code for
reference.
4.1 UMAP for screening results
example:
Suppose you have performed all algorithm screening on your Seurat object and wish to examine the distribution across different celltypes and patient, you may reference and use the following code:
library(zeallot)
# library(Seurat)
# library(patchwork)
# library(purrr)
c(
celltype_umap,
patient_umap,
scissor_umap,
scab_umap,
scpas_umap,
scpp_umap,
degas_umap
) %<-%
purrr::map(
# make sure these column names exist
c("celltype", "patient", "scissor", "scAB", "scPAS", "scPP", "DEGAS"),
~ Seurat::DimPlot(
your_seurat_obj,
group.by = .x,
pt.size = 0.05,
reduction = "umap"
) +
ggplot2::ggtitle(.x)
)
# * Show
umaps = celltype_umap +
patient_umap +
scissor_umap +
scab_umap +
scpas_umap +
scpp_umap +
degas_umap +
patchwork::plot_layout(ncol = 3)
umapsThis will generate seven UMAP plots separately.
Or suppose you have performed scPAS screening on your
Seurat object and want to visualize the distribution of prediction
confidence scores, you may reference and use the following code:
library(zeallot)
library(patchwork)
# library(Seurat)
# library(purrr)
c(scPAS_Pvalue_umap, scPAS_NRS_umap) %<-%
purrr::map(
c("scPAS_Pvalue", "scPAS_NRS"),
~ Seurat::FeaturePlot(
object = your_seurat_obj,
features = .x,
) +
ggplot2::ggtitle(.x) +
theme(legend.position = "right")
)
# * Show
scPAS_Pvalue_umap | scPAS_NRS_umapThis will generate two plots, one for each feature specified in
feature.
helpful documentation:
Seurat::DimPlot - https://satijalab.org/seurat/reference/DimPlot.html
Seurat::FeaturePlot - https://satijalab.org/seurat/reference/FeaturePlot.html
4.2 Stack bar plot for screening results
Key parameters for ScreenFractionPlot:
-
seurat_obj: A Seurat object after screening. -
group_by: Used to specify the column of the meta.data inseurat_obj. The plot results will be grouped by this parameter. -
screen_type: Screening algorithm used before. (case-sensitive, e.g., “scissor” for Scissor results) -
show_null: Logical, whether to show groups with zero cells (default: FALSE). -
plot_colorCustom color palette (named vector format):- Required names: “Positive”, “Negative”, “Neutral”, “Other”
- Default: c(“Neutral”=“#CECECE”, “Other”=“#CECECE”, “Positive”=“#ff3333”, “Negative”=“#386c9b”)
Suppose you have already performed the scPAS algorithm
screening on your Seurat object, and you want to check the proportion of
positive cells across different patients. You can refer to and use the
following code:
plot <- ScreenFractionPlot(
screened_seurat = scpas_result$scRNA_data,
group_by = "patient", # grouping basis for the x-axis
screen_type = "scPAS",
plot_title = "scPAS Screening Results"
)If you have performed multiple screening methods and already merged the results, you can use the following code:
plot <- ScreenFractionPlot(
screened_seurat = merged_seurat,
group_by = "patient", # grouping basis for the x-axis
screen_type = c("scPAS", "scAB", "scPP"), # multiple screening results
plot_title = "Screening Results" # A screen_type prefix will be added to the current plot title
)The order of the groups is determined by the proportion of Positive cells within each group.
returning structure:
If a single screen_type is specified
-
stats: A data frame containing the proportion of positive cells for each group. -
plot: A ggplot2 object.
If multiple screen_types are specified
-
stats: A list containing data frames containing the proportion of positive cells for each group. -
plot: A list containing each ggplot2 objects. -
combined_plot: A ggplot2 object containing all the plots (2*2 grid).
4.3 Venn diagram for screening results
ggVennDiagram is used to generate a Venn diagram for the
screening results. Suppose you have performed some of the screening
algorithms on your Seurat object, and you want to check the overlap of
the cells selected by each algorithm. You can refer to and use the
following code:
example:
library(ggVennDiagram)
# # *If you have merged the results, you can use the following code instead:
# c(scissor_pos, scab_pos, scpas_pos, scpp_pos, degas_pos) %<-%
# purrr::map(
# c("scissor", "scAB", "scPAS", "scPP", "DEGAS"),
# ~ colnames(merged_seurat)[
# which(merged_seurat[[.x]] == "Positive")
# ]
# )
# * get the cell vectors
scissor_pos <- colnames(scissor_result$scRNA_data)[
which(scissor_result$scRNA_data$scissor == "Positive")
]
scab_pos <- colnames(scab_result$scRNA_data)[
which(scab_result$scRNA_data$scAB == "Positive")
]
scpas_pos <- colnames(scpas_result$scRNA_data)[
which(scpas_result$scRNA_data$scPAS == "Positive")
]
scpp_pos <- colnames(scissor_result$scRNA_data)[
which(scpp_result$scRNA_data$scPP == "Positive")
]
degas_pos <- colnames(degas_result$scRNA_data)[
which(degas_result$scRNA_data$DEGAS == "Positive")
]
all_cells <- colnames(your_seurat_obj)
# * create a list of cell vectors
pos_venn = list(
scissor = scissor_pos,
scpas = scpas_pos,
scab = scab_pos,
scpp = scpp_pos,
degas = degas_pos,
all_cells = all_cells
# * you can add more groups here
)
set.seed(123)
venn_plot = ggVennDiagram::ggVennDiagram(
x = pos_venn,
# * the labels of each group to be shown on the diagram
category.names = c(
"Scissor",
"scPAS",
"scAB",
"scPP",
"DEGAS",
"All cells"
),
# * the colors of each group
set_color = c(
"#a33333ff",
"#37ae00ff",
"#2a2a94ff",
"#9c8200ff",
"#bb14adff",
"#008383ff"
)
) +
ggplot2::scale_fill_gradient(low = "white", high = "#ffb6b6ff") +
ggplot2::ggtitle("Screening Venn Diagram")helpful link:
4.4 Upset plot for screening results
If too many screening meyhods are selected, the number of intersections among cells screened by different methods will also increase. In this case, using an upset plot is more intuitive and neat than a Venn diagram.
ggupset is used for visualizing upset plot.
Key parameters for ScreenUpset:
-
screened_seurat: A Seurat object after screening. -
screen_type: Screening algorithm used before. (case-sensitive, e.g., “scissor” for Scissor results) -
n_intersections: Number of intersections to display in the plot. Default: 20. -
x_lab: Label for the x-axis. Default: “Screen Set Intersections”. -
y_lab: Label for the y-axis. Default: “Number of Cells”. -
title: Plot title. Default: “Cell Counts Across Screen Set Intersections”. -
bar_color: Color for the bars in the plot. Default: “#4E79A7”. -
combmatrix_point_color: Color for points in the combination matrix. Default: “black”. -
...: Additional arguments passed toggplot2::theme()for customizing the plot appearance.
upset <- ScreenUpset(screened_seurat = merged_seurat)returning structure:
-
plot: A ggplot2 object. -
stats: A data frame containing the numbers of positive cells for each group.
helpful link:
ggupset - https://github.com/const-ae/ggupset
5. Example
5.1 Survival-associated cell screening
Here we use the example data LUAD to demonstrate how to use the
functions in SigBridgeR to screen cells associated with
phenotype.
# Set working directory
if (requireNamespace("here", quietly = TRUE)) {
setwd(here::here())
knitr::opts_knit$set(root.dir = here::here())
}
library(SigBridgeR)
library(zeallot)
library(Seurat)
# * load the example data
c(mat_exam, bulk, pheno) %<-% LoadRefData(data_type = "survival")
dim(mat_exam)
#[1] 33694 1093
dim(bulk)
# [1] 4071 506
bulk[1:6,1:6]
# TCGA-69-7978 TCGA-62-8399 TCGA-78-7539 TCGA-73-4658 TCGA-44-6775 TCGA-44-2655
# HIF3A 4.2598 11.6239 9.1362 5.0288 4.0573 5.5335
# RTN4RL2 8.2023 5.5819 3.5365 7.4156 7.7107 5.3257
# HMGCLL1 2.7476 5.8513 3.8334 3.6447 2.9188 4.8820
# LRRTM1 0.0000 0.4628 4.7506 6.8005 7.7819 2.2882
# GRIN1 6.6074 5.4257 4.9563 7.3510 3.5361 3.3311
# LRRTM3 1.7458 2.0092 0.0000 1.4468 0.0000 0.0000
nrow(pheno)
# [1] 506
head(pheno)
# time status
# TCGA-69-7978 4.40 0
# TCGA-62-8399 88.57 0
# TCGA-78-7539 25.99 0
# TCGA-73-4658 52.56 1
# TCGA-44-6775 23.16 0
# TCGA-44-2655 43.50 0This single-cell RNA data is from humans. We set many parameters to
FALSE or 0 in order to maximize the
flexibility of downstream analyses and capture a broader range of
biological signals, so as to avoid insignificant results caused by too
small a dataset.
Now we use SCPreProcess() to pre-process the single-cell
RNA expression matrix data.
seurat = SCPreProcess(
sc = mat_exam,
quality_control = FALSE,
data_filter = FALSE,
min_cells = 0,
min_features = 0,
scale_features = rownames(mat_exam),
dims = 1:20,
resolution = 0.1
)BulkPreProcess() is to pre-process the bulk expression
data and the related clinical data.frame (containing bulk sample
information) to perform PCA. Here, we want to retain the data so that
the screening can reflect the most accurate situation. You can choose
whether to filter or retain based on your own needs. See also 2.2 Bulk expression data for more
details.
To facilitate the demonstration of the use of the sample_info
parameter, we will divide the pheno based on its survival
status into two groups, which will be stored in the
condition column.
sample_info <- tibble::rownames_to_column(pheno, var = "sample") %>%
dplyr::rename(condition = status)
head(sample_info)
# sample time condition
# 1 TCGA-69-7978 4.40 0
# 2 TCGA-62-8399 88.57 0
# 3 TCGA-78-7539 25.99 0
# 4 TCGA-73-4658 52.56 1
# 5 TCGA-44-6775 23.16 0
# 6 TCGA-44-2655 43.50 0
bulk = BulkPreProcess(
data = bulk,
sample_info = sample_info,
min_count_threshold = 0,
min_gene_expressed = 0,
min_total_reads = 0,
min_genes_detected = 0,
min_correlation = 0.1
)
# ℹ [2025/10/11 09:17:46] Starting data preprocessing...
# ✔ [2025/10/11 09:17:47] Data loaded: 4071 genes * 506 samples
# ℹ [2025/10/11 09:17:47] Computing basic statistics...
# ✔ [2025/10/11 09:17:47] 4071 genes pass expression filter
# ℹ [2025/10/11 09:17:47] Starting detailed quality checks...
# ℹ [2025/10/11 09:17:47] Computing sample correlations...
# ✔ [2025/10/11 09:17:49] Correlation analysis completed: min correlation = 0.288
# ℹ [2025/10/11 09:17:49] Performing principal component analysis...
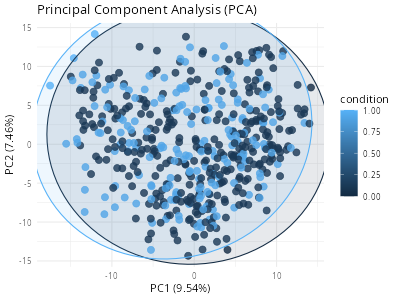
# ℹ [2025/10/11 09:17:49] PCA completed: PC1(9.54%) PC2(7.46%), 7 outlier samples
# Warning: Outlier samples detected: TCGA-49-6742, TCGA-55-8513, TCGA-55-8094, TCGA-78-7150, TCGA-78-7220, TCGA-MP-A4TA, TCGA-50-5072
# ℹ [2025/10/11 09:17:49] Generating visualization plots...
# ✔ [2025/10/11 09:17:49] Plot generation completed
# ℹ [2025/10/11 09:17:50] Performing data filtering...
# ✔ [2025/10/11 09:17:50] Data filtering completed:
# ℹ Genes: 4071 -> 4071 (removed 0)
# ℹ Samples: 506 -> 506 (removed 0)
# ℹ Quality Assessment Summary:
# ✔ Data integrity: No missing values
# ✔ Sample read depth: All samples pass threshold (>=0)
# ✔ Gene detection: All samples pass threshold (>=0)
# ✔ Sample correlation: Good (minimum = 0.288)
# Warning: Sample outliers: 7 sample(s) detected
# ✔ [2025/10/11 09:17:50] BulkPreProcess completedAnd we will see a PCA plot.
knitr::include_graphics("vignettes/example_figures/bulk_preprocess_pca.png")Thus far, we have completed all the data preprocessing. We are now ready to formally employ various single-cell phenotypic screening algorithms.
First, we use scissor to screen cells associated with
survival.
scissor_result <- Screen(
matched_bulk = bulk,
sc_data = seurat,
phenotype = pheno,
label_type = "survival",
phenotype_class = "survival",
screen_method = "Scissor",
alpha = 0.05
)
# ℹ [2025/09/08 17:03:20] Scissor start...
# ℹ [2025/09/08 17:03:20] Start from raw data...
# ℹ Using "RNA_snn" graph for network.
# ℹ [2025/09/08 17:03:20] Normalizing quantiles of data...
# ℹ [2025/09/08 17:03:20] Subsetting data...
# ℹ [2025/09/08 17:03:21] Calculating correlation...
# ----------------------------------------------------------------------------------------------------
# Five-number summary of correlations:
# 0% 25% 50% 75% 100%
# -0.2342323 0.0594385 0.1118708 0.1642065 0.5250605
# ----------------------------------------------------------------------------------------------------
# ℹ [2025/09/08 17:03:21] Perform cox regression on the given clinical outcomes:
# ✔ Statistics data saved to `Scissor_inputs.RData`.
# ℹ [2025/09/08 17:03:22] Screening...
#
# ── At alpha = 0.05 ──
#
# Scissor identified 265 Scissor+ cells and 73 Scissor- cells.
# The percentage of selected cell is: 30.924%
table(scissor_result$scRNA_data$scissor)
# Negative Neutral Positive
# 73 755 265 You will see an additional “Scissor_inputs.RData” in the working
directory. This is the intermediate data generated by the Scissor
method, which we can use to save running time ( You can also set
path2load_scissor_cache=NULL to suppress the saving of
intermediate files ). Meanwhile, we set
reliability_test=TRUE, which will run an additional
reliability test.
scissor_result <- Screen(
matched_bulk = bulk, # doesn't need to be provided Since the intermediate data is provided
sc_data = seurat,
phenotype = pheno_ok, # doesn't need to be provided Since the intermediate data is provided
label_type = "survival",
phenotype_class = "survival",
screen_method = "Scissor",
alpha = 0.05,
path2load_scissor_cache = "Scissor_inputs.RData",
reliability_test = TRUE
)
# ℹ [2025/09/08 16:07:48] Scissor start...
# ℹ [2025/09/08 16:07:48] Loading data from `Scissor_inputs.RData`...
# ℹ [2025/09/08 16:07:48] Screening...
# [1] "alpha = 0.05"
# [1] "Scissor identified 249 Scissor+ cells and 245 Scissor- cells."
# [1] "The percentage of selected cell is: 45.197%"
# --------------------------------------------------------------------------------
# ℹ [2025/09/08 16:07:54] Start reliability test
# Attaching package: ‘survival’
# The following object is masked from ‘package:future’:
# cluster
# [1] "|**************************************************|"
# [1] "Perform cross-validation on X with true label"
# Finished!
# [1] "|**************************************************|"
# [1] "Perform cross-validation on X with permutated label"
# Finished!
# [1] "Test statistic = 0.590"
# [1] "Reliability significance test p = 0.000"
# ✔ [2025/09/08 16:10:54] reliability test: Done
scissor_result$reliability_result$statistic
# [1] 0.5899587
scissor_result$reliability_result$p
# [1] 0
scissor_result$reliability_result$c_index_test_real
# [1] 0.6038544 0.5022222 0.6050725 0.6279391 0.5064935 0.6033520 0.6769231 0.6453089 0.4968421 0.6315789
scissor_result$reliability_result$c_index_test_back %>% unlist() %>% matrix(nrow = 10)
# [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
# [1,] 0.5594714 0.4943820 0.5379747 0.5583658 0.5527728 0.6146435 0.5130597 0.5701275 0.4691943 0.5177305
# [2,] 0.5735608 0.4726891 0.4146341 0.5020661 0.5354331 0.6840149 0.5540275 0.4990758 0.5324484 0.5497382
# [3,] 0.5960145 0.5157116 0.6191446 0.5091912 0.5659656 0.5870370 0.5090580 0.4247788 0.5734266 0.5539715
# [4,] 0.6210191 0.6563147 0.5581948 0.4675615 0.4688222 0.5308219 0.5100402 0.6709091 0.6155779 0.5607143
# [5,] 0.4623656 0.5716695 0.4920441 0.5403727 0.5555556 0.5911215 0.5738499 0.6189258 0.5871560 0.6084337
# [6,] 0.5868794 0.5127660 0.7416880 0.5366876 0.5417440 0.5633803 0.5161943 0.4830372 0.4732965 0.5740132
# [7,] 0.4975610 0.5751503 0.4801902 0.5588972 0.4940898 0.5723370 0.5788382 0.6171875 0.5884413 0.5445545
# [8,] 0.6331361 0.5970516 0.5473888 0.6274131 0.5159705 0.5000000 0.5299760 0.4905660 0.5277778 0.5027322
# [9,] 0.6390805 0.6548913 0.5049310 0.6299639 0.6385135 0.5964392 0.5381605 0.7462687 0.5185185 0.6585859
# [10,] 0.5207373 0.5000000 0.6051780 0.4932127 0.6892430 0.4786517 0.5619266 0.6614583 0.6502242 0.5333333Next, we use scPAS, scAB and scPP to do the same screening. Generally
you only need to change the screen_method, as long as you
have not specified any particular parameters.
scpas_result <- Screen(
matched_bulk = bulk,
sc_data = seurat,
phenotype = pheno,
label_type = "survival",
phenotype_class = "survival",
screen_method = "scPAS",
alpha = 0.05
)
# ℹ [2025/10/20 16:24:27] Start scPAS screening.
# ℹ [2025/10/20 16:24:29] Quantile normalization of bulk data.
# ℹ [2025/10/20 16:24:29] Extracting single-cell expression profiles...
# ℹ [2025/10/20 16:24:29] Constructing a gene-gene similarity by single cell data...
# Building SNN based on a provided distance matrix
# Computing SNN
# ℹ [2025/10/20 16:24:30] Optimizing the network-regularized sparse regression model...
# ℹ [2025/10/20 16:24:30] Perform cox regression on the given phenotypes...
#
# ── At alpha = 0.05 ──
#
# lambda = 0.776168989003421
# scPAS identified 59 risk+ features and 61 risk- features.
# The percentage of selected feature is: 13.73%
# ℹ [2025/10/20 16:24:49] Calculating quantified risk scores...
# ℹ [2025/10/20 16:24:49] Qualitative identification by permutation test program with 2000 times random perturbations...
# ✔ [2025/10/20 16:24:50] scPAS screening done.
table(scpas_result$scRNA_data$scPAS)
# Negative Neutral Positive
# 5 1085 3 As you can see, due to differences in data and algorithms, not every
screening algorithm is able to screen out cells. You can adjust the
corresponding parameters, e.g. change the alpha to
NULL, this will make scPAS iterate alpha until the result
is significant (or judged as having no significant cell subpopulations).
See also 3.2 (Option B) scPAS
Screening
scpas_result <- Screen(
matched_bulk = bulk,
sc_data = seurat,
phenotype = pheno,
label_type = "survival",
phenotype_class = "survival",
screen_method = "scPAS",
alpha = NULL,
cutoff = 0.2
)
# ℹ [2025/10/20 16:43:31] Start scPAS screening.
# ℹ [2025/10/20 16:43:32] Quantile normalization of bulk data.
# ℹ [2025/10/20 16:43:32] Extracting single-cell expression profiles...
# ℹ [2025/10/20 16:43:32] Constructing a gene-gene similarity by single cell data...
# Building SNN based on a provided distance matrix
# Computing SNN
# ℹ [2025/10/20 16:43:33] Optimizing the network-regularized sparse regression model...
# ℹ [2025/10/20 16:43:33] Perform cox regression on the given phenotypes...
#
# ── At alpha = 0.001 ──
#
# lambda = 7.61825638357188
# scPAS identified 315 risk+ features and 366 risk- features.
# The percentage of selected feature is: 77.918%
#
# ── At alpha = 0.005 ──
#
# lambda = 3.68743152046651
# scPAS identified 193 risk+ features and 231 risk- features.
# The percentage of selected feature is: 48.513%
#
# ── At alpha = 0.01 ──
#
# lambda = 2.55333682907113
# scPAS identified 137 risk+ features and 158 risk- features.
# The percentage of selected feature is: 33.753%
#
# ── At alpha = 0.05 ──
#
# lambda = 0.776168989003421
# scPAS identified 59 risk+ features and 61 risk- features.
# The percentage of selected feature is: 13.73%
# ℹ [2025/10/20 16:44:55] Calculating quantified risk scores...
# ℹ [2025/10/20 16:44:55] Qualitative identification by permutation test program with 2000 times random perturbations...
# ✔ [2025/10/20 16:44:57] scPAS screening done.Generally speaking, a larger alpha means looser screening. However,
it still won’t result in the occurence of too many false-positive cells.
You can also adjust the cutoff parameter as you like.
table(scpas_result$scRNA_data$scPAS)
# Negative Neutral Positive
# 5 1085 3 Now we use scAB, scPP and DEGAS to screen cells.
scab_result = Screen(
matched_bulk = bulk,
sc_data = seurat,
phenotype = pheno,
label_type = "survival",
phenotype_class = "survival",
screen_method = "scAB"
)
# ℹ [2025/09/08 17:04:51] Start scAB screening.
# ℹ Using "RNA_snn" graph for network.
# ℹ [2025/09/08 17:04:52] Selecting K...
# ℹ [2025/09/08 17:06:15] Run NMF with phenotype and cell-cell similarity regularization at K = 3.
# ℹ [2025/09/08 17:06:19] Screening cells...
# ℹ [2025/09/08 17:06:19] scAB screening done.
table(scab_result$scRNA_data$scAB)
# Other Positive
# 1018 75
scpp_result = Screen(
matched_bulk = bulk,
sc_data = seurat,
phenotype = pheno,
label_type = "survival",
phenotype_class = "survival",
screen_method = "scPP"
)
# ℹ [2025/09/08 17:00:28] Start scPP screening.
# ℹ [2025/09/08 17:00:28] Finding markers...
# Warning in coxph.fit(X, Y, istrat, offset, init, control, weights = weights, :
# Loglik converged before variable 1 ; coefficient may be infinite.
# ℹ [2025/09/08 17:00:52] Screening...
# Genes in the gene sets NOT available in the dataset:
# gene_pos: 13 (6% of 230)
# gene_neg: 54 (12% of 446)
# There are no genes significantly upregulated in Phenotype- compared to Phenotype+.
# ✔ [2025/09/08 17:00:54] scPP screening done.
table(scpp_result$scRNA_data$scPP)
# Negative Neutral Positive
# 52 993 48
# I recommend running this code in the background.
degas_result = Screen(
matched_bulk = bulk,
sc_data = seurat,
phenotype = pheno,
label_type = "This_is_a_DEGAS_test",
phenotype_class = "survival",
screen_method = "DEGAS"
)
# ℹ [2025/10/09 18:41:00] Starting DEGAS Screen
# ℹ [2025/10/09 18:41:01] Setting up Environment...
# ℹ [2025/10/09 18:41:08] Training DEGAS model...
# ℹ [2025/10/09 18:41:08] 3-layer DenseNet BlankCox DEGAS model
# ℹ [2025/10/09 18:41:10] Python check passed, using Python 3.9.15
# ℹ [2025/10/09 18:41:10] Training...
######################
# Many python output #
######################
# ℹ [2025/10/10 17:35:04] Predicting and Labeling...
# ℹ [2025/10/10 17:35:04] Labeling screened cells...
# ℹ [2025/10/10 17:35:04] Searching for survival-associated cells...
# ℹ Scores over 0.499 are considered `Positive`.
# ℹ [2025/10/10 17:35:04] DEGAS Screen done.
table(degas_result$scRNA_data$DEGAS)
# Other Positive
# 1038 55 After these algorithms have been run, the four sets of data can be merged since screening methods performed on the same data.
screen_result = MergeResult(
scissor_result,
scpas_result,
scab_result,
scpp_result,
degas_result
)
# ✔ Successfully merged 5 objects.
class(screen_result)
# [1] "Seurat"
# attr(,"package")
# [1] "SeuratObject"
colnames(screen_result[[]])
# [1] "orig.ident" "nCount_RNA" "nFeature_RNA" "RNA_snn_res.0.1" "seurat_clusters" "scissor" "scPAS_RS" "scPAS_NRS" "scPAS_Pvalue" "scPAS_FDR"
# [11] "scPAS" "scAB" "scAB_Subset1" "Subset1_loading" "scAB_Subset2" "Subset2_loading" "scPP_AUCup" "scPP_AUCdown" "scPP" "DEGAS" Finally, we can visualize the screening results. Let’s start with a Venn diagram to see the situation.
library(ggVennDiagram)
library(zeallot)
# * color palette
set.seed(123)
my_colors <- randomcoloR::distinctColorPalette(
length(unique(screen_result$seurat_clusters)),
runTsne = TRUE
)
c(scissor_pos, scab_pos, scpas_pos, scpp_pos, degas_pos) %<-%
purrr::map(
c("scissor", "scAB", "scPAS", "scPP", "DEGAS"),
~ colnames(screen_result)[
which(screen_result[[.x]] == "Positive")
]
)
all_cells <- colnames(screen_result)
# * create a list of cell vectors
pos_venn = list(
scissor = scissor_pos,
scpas = scpas_pos,
scab = scab_pos,
scpp = scpp_pos,
degas = degas_pos,
all_cells = all_cells
)
set.seed(123)
venn = ggVennDiagram::ggVennDiagram(
x = pos_venn,
# * the labels of each group to be shown on the diagram
category.names = c(
"Scissor",
"scPAS",
"scAB",
"scPP",
"DEGAS",
"All cells"
),
# * the colors of each group
set_color = c(
"#a33333ff",
"#37ae00ff",
"#0000f5ff",
"#d4b100ff",
"#e600eeff",
"#008383ff"
),
label_geom = "text"
) +
ggplot2::scale_fill_gradient(low = "white", high = "#ffb6b6ff") +
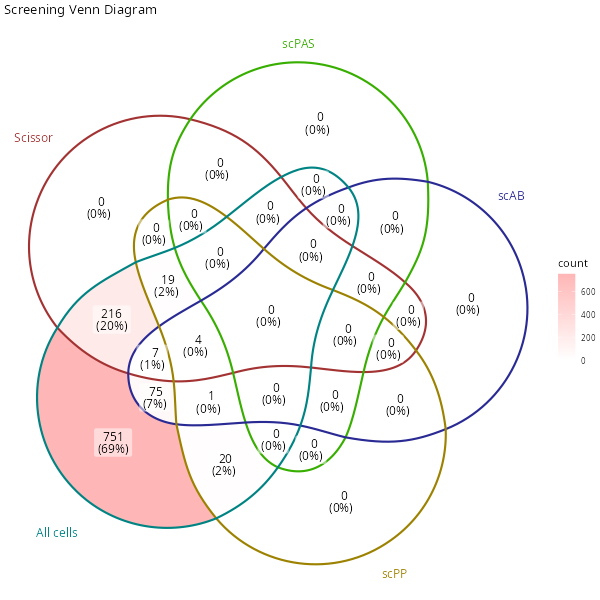
ggplot2::ggtitle("Screening Venn Diagram")
venn
# ggplot2::ggsave(
# "vignettes/example_figures/venn.png",
# plot = venn,
# width = 10,
# height = 10
# )
knitr::include_graphics("vignettes/example_figures/venn.png")When there are too many sets, the visualization effect of the Venn diagram is not very good. We can use a set plot instead. Only positive cells are shown in the set plot.
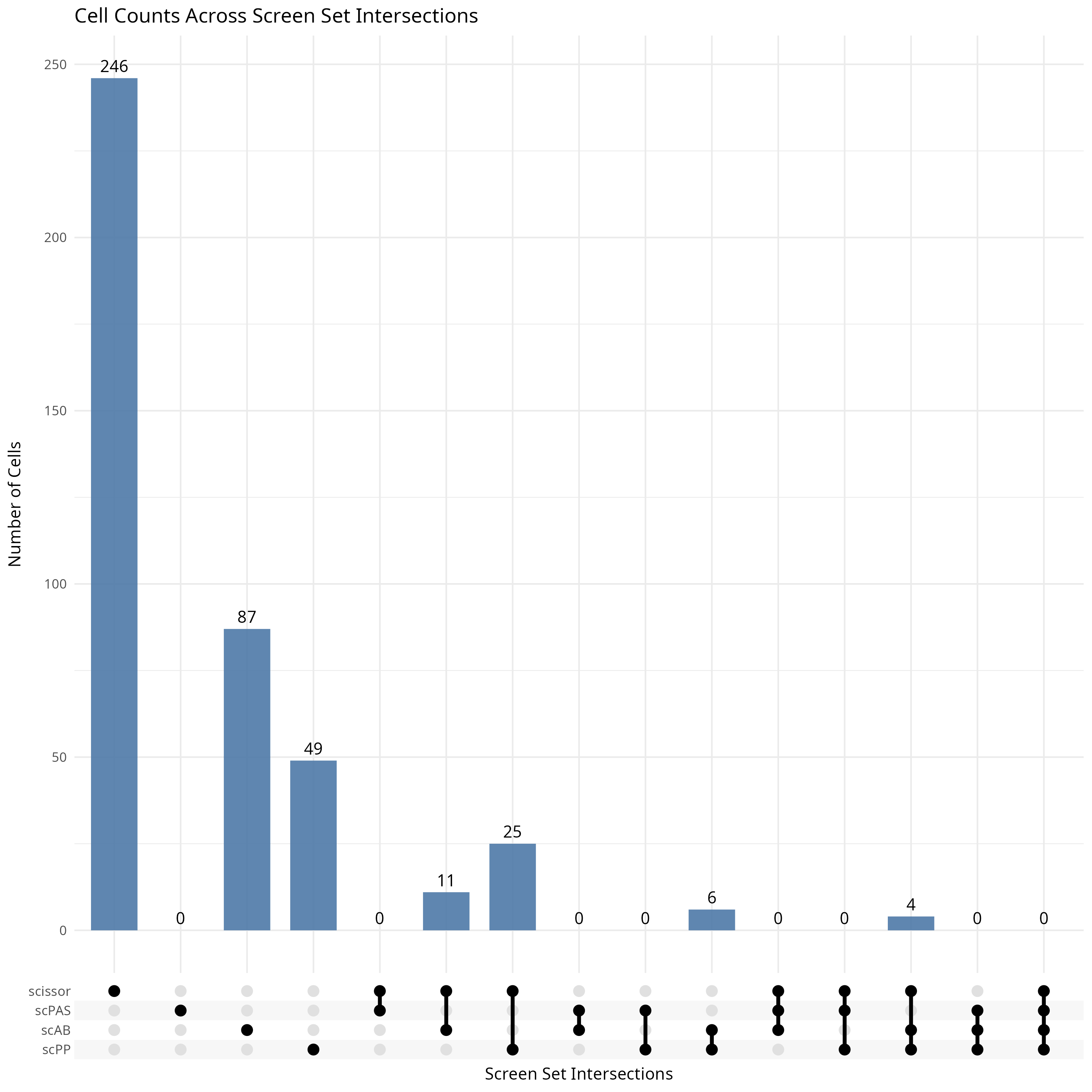
upset <- ScreenUpset(
screened_seurat = screen_result,
screen_type = c("scissor", "scPAS", "scAB", "scPP", "DEGAS"),
n_intersections = 40
)
# * show the cell numbers of each set
head(upset$stats)
# # A tibble: 6 × 3
# intersection sets count
# <chr> <named list> <dbl>
# 1 scissor <chr [1]> 265
# 2 scPAS <chr [1]> 2
# 3 scAB <chr [1]> 75
# 4 scPP <chr [1]> 49
# 5 DEGAS <chr [1]> 55
# 6 scissor & scPAS <chr [2]> 1
# ggplot2::ggsave(
# "vignettes/example_figures/upset.png",
# plot = upset$plot,
# width = 10,
# height = 10
# )
knitr::include_graphics("vignettes/example_figures/upset.png")A bar chart showing proportions can also be used to examine the
screening results. Since the example data does not have sample metadata,
we have created a fictional Sample column.
set.seed(123)
# *fictional sample column
screen_result$Sample <- sample(
paste0("Sample", 1:10),
ncol(screen_result),
replace = TRUE
)
table(screen_result$Sample)
# Sample1 Sample10 Sample2 Sample3 Sample4 Sample5 Sample6 Sample7 Sample8 Sample9
# 98 117 96 119 95 100 101 131 115 121
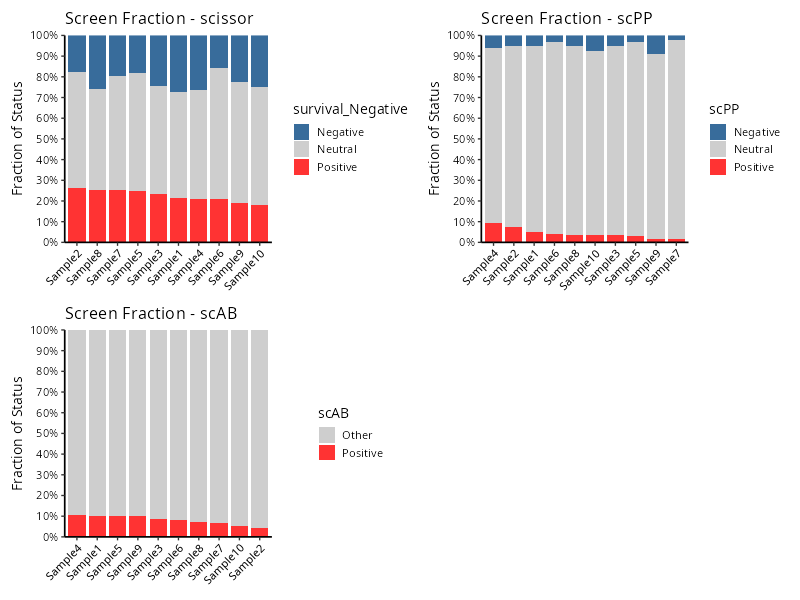
fraction_list = ScreenFractionPlot(
screened_seurat = screen_result,
group_by = "Sample",
screen_type = c("scissor", "scPP", "scAB", "scPAS", "DEGAS"),
show_null = FALSE,
plot_color = NULL,
show_plot = TRUE
)
# Creating plots for 5 screen types...
# ggplot2::ggsave(
# "vignettes/example_figures/fraction.png",
# plot = fraction_list$combined_plot,
# width = 10,
# height = 10
# )
knitr::include_graphics("vignettes/example_figures/fraction.png")As you see, the label_types set in function
Screen are shown in the legend of each plot.
The fraction_list contains the statistical data and
charts for each screening algorithm. The title of each plot will be
appended with the method name as an identifier.
fraction_list
├── stats
│ ├── scissor
│ ├── scAB
│ ├── scPAS
│ ├── scPP
│ └── DEGAS
├── plots
│ ├── scissor
│ ├── scAB
│ ├── scPAS
│ ├── scPP
│ └── DEGAS
└── combined_plot # show 5 plots in one plotUMAP is the most commonly used type of plot in academic literature.
library(patchwork)
library(zeallot)
my_palette <- randomcoloR::distinctColorPalette(
k = length(unique(screen_result$Sample)),
runTsne = TRUE
)
sample_umap = Seurat::DimPlot(
screen_result,
group.by = "Sample",
pt.size = 1.2,
alpha = 0.8,
reduction = "umap",
cols = my_palette
) +
ggplot2::ggtitle("Sample")
c(
scissor_umap,
scab_umap,
scpas_umap,
scpp_umap,
degas_umap
) %<-%
purrr::map(
c("scissor", "scAB", "scPAS", "scPP", "DEGAS"), # make sure these column names exist
~ Seurat::DimPlot(
screen_result,
group.by = .x,
pt.size = 1.2,
alpha = 0.8,
reduction = "umap",
cols = c(
"Neutral" = "#CECECE",
"Other" = "#CECECE",
"Positive" = "#ff3333",
"Negative" = "#386c9b"
)
) +
ggplot2::ggtitle(.x)
)
# * Show
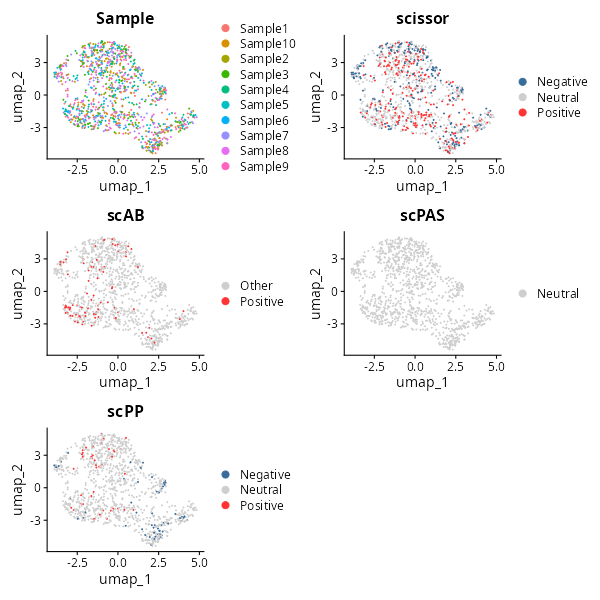
umaps = sample_umap +
scissor_umap +
scab_umap +
scpas_umap +
scpp_umap +
degas_umap +
plot_layout(ncol = 2)
umaps
# ggsave(
# "vignettes/example_figures/umaps.png",
# plot = umaps,
# width = 10,
# height = 10
# )
knitr::include_graphics("vignettes/example_figures/umaps.png")All the analysis has been completed, and we can save the data for future use.
SeuratObject::SaveSeuratRds(object = screen_result, filename = "screened_result.rds")5.2 Continuous phenotype associated cell screening
Generally speaking, the process is the same as described in 5.1 Survival-associated cell screening. Here, only the preprocessing of continuous phenotypic data is introduced.
library(SigBridgeR)
library(zeallot)
library(Seurat)
setwd(here::here())
# * load the example data
c(mat_exam, bulk, pheno) %<-% LoadRefData(data_type = "continuous")
dim(mat_exam)
#[1] 33694 1093
dim(bulk)
# [1] 4106 289
bulk[1:6,1:6]
# TCGA-AZ-6599-01 TCGA-AA-3655-01 TCGA-A6-6137-01 TCGA-CK-4952-01 TCGA-A6-5657-01 TCGA-AD-6963-01
# HIF3A 2.3437 2.0858 6.0759 1.9506 5.4777 4.4634
# CAMK4 4.9331 2.3709 4.1387 1.1557 4.1746 3.2363
# RNF112 2.4817 2.4947 3.5941 2.3486 4.9185 1.4621
# SPN 5.6704 6.8577 8.0598 5.0049 7.6076 7.3960
# LRRTM1 1.6031 0.9465 1.9142 0.0000 3.2523 0.0000
# GRIN1 6.4944 4.3225 2.8073 7.3460 4.5000 3.1816
# * A named vector
head(pheno)
# TCGA-AZ-6599-01 TCGA-AA-3655-01 TCGA-A6-6137-01 TCGA-CK-4952-01 TCGA-A6-5657-01 TCGA-AD-6963-01
# 178 65 91 206 63 67 If your phenotype is a data.frame, try this to convert
it to a named vector:
pheno = setNames(your_data.frame$continuous_numeric, your_data.frame$sample)5.3 Binarized phenotype associated cell screening
This process is also the same as described in 5.1 Survival-associated cell screening. Here, only the preprocessing of binary phenotypic data is introduced.
library(SigBridgeR)
library(zeallot)
library(Seurat)
setwd(here::here())
# * load the example data
c(mat_exam, bulk, pheno) %<-% LoadRefData(data_type = "binary")
dim(mat_exam)
#[1] 33694 1093
dim(bulk)
# [1] 4106 434
bulk[1:6,1:6]
# TCGA-CA-5256-01 TCGA-AZ-6599-01 TCGA-AA-3655-01 TCGA-A6-6137-01 TCGA-CK-4952-01 TCGA-A6-5657-01
# HIF3A 3.7172 2.3437 2.0858 6.0759 1.9506 5.4777
# CAMK4 3.0698 4.9331 2.3709 4.1387 1.1557 4.1746
# RNF112 1.3702 2.4817 2.4947 3.5941 2.3486 4.9185
# SPN 5.5207 5.6704 6.8577 8.0598 5.0049 7.6076
# LRRTM1 3.2408 1.6031 0.9465 1.9142 0.0000 3.2523
# GRIN1 3.0698 6.4944 4.3225 2.8073 7.3460 4.5000
# * A named vector
head(pheno)
# TCGA-CA-5256-01 TCGA-AZ-6599-01 TCGA-AA-3655-01 TCGA-A6-6137-01 TCGA-CK-4952-01 TCGA-A6-5657-01
# 1 1 1 1 1 1 If your phenotype is a data.frame, your binary variable
is stored in the “data” column, categorized as ‘Tumor’ and ‘Normal’, we
will assign ‘Tumor’ a value of 1 and ‘Normal’ a value of 0. In this way,
cells screened as Positive will be associated with
‘Tumor’. Try this to convert it to a named vector:
pheno = mutate(
pheno,
data = dplyr::case_when(
data == "Tumor" ~ 1,
data == "Normal" ~ 0
)
)
pheno = setNames(pheno$data, pheno$Sample)6. Other function details
6.1 Add miscellaneous information to the Seurat object
SigBridgeR uses AddMisc() to record what data features
or evidence the various screening algorithms are based on during
execution.
-
AddMisc(): Add miscellaneous information to the Seurat object. Support for adding multiple attributes to theSeuratObject@miscslot simultaneously.
# basic usage
seurat_obj <- AddMisc(seurat_obj, "QC_stats" = qc_df)
# Auto-incrementing example when `cover` set to FALSE
seurat_obj <- AddMisc(seurat_obj, markers = markers1)
seurat_obj <- AddMisc(seurat_obj, markers = markers2, cover=FALSE)
# Add multiple attributes to the `SeuratObject@misc` slot simultaneously
seurat_obj <- AddMisc(seurat_obj, markers1 = markers1, markers2 = markers2)6.2 Load reference data
Parameters
-
data_type: The type of data to load. Can be either “continuous”, “survival” or “binary”, case-insensitive. -
path: The path to the data directory. -
cache: Whether to cache the data. Defaults toTRUE. -
timeout: The maximum timeout time when downloading data.
When loading the example data, the single-cell RNA expression matrix, the bulk RNA expression matrix, and the clinical phenotype data are loaded all at once. These data are combined into a list and returned.
mydata <- LoadRefData(
data_type = c("survival"),
path = tempdir(),
cache = TRUE,
timeout = 60
)
# * mat_exam (matrix_example)
mydata[[1]][1:6,1:6]
# SMC01.T_AAACCTGCATACGCCG SMC01.T_AAACCTGGTCGCATAT SMC01.T_AAACCTGTCCCTTGCA SMC01.T_AAACGGGAGGGAAACA SMC01.T_AAACGGGGTATAGGTA SMC01.T_AAAGATGAGGCCGAAT
# A1BG 0 0 0 0 0 0
# A1BG.AS1 0 0 0 0 0 0
# A1CF 0 2 0 0 3 0
# A2M 0 0 0 0 0 0
# A2M.AS1 0 0 0 0 0 0
# A2ML1 0 0 0 0 0 0
# * bulk_survival
mydata[[2]][1:6,1:6]
# TCGA-69-7978 TCGA-62-8399 TCGA-78-7539 TCGA-73-4658 TCGA-44-6775 TCGA-44-2655
# HIF3A 4.2598 11.6239 9.1362 5.0288 4.0573 5.5335
# RTN4RL2 8.2023 5.5819 3.5365 7.4156 7.7107 5.3257
# HMGCLL1 2.7476 5.8513 3.8334 3.6447 2.9188 4.8820
# LRRTM1 0.0000 0.4628 4.7506 6.8005 7.7819 2.2882
# GRIN1 6.6074 5.4257 4.9563 7.3510 3.5361 3.3311
# LRRTM3 1.7458 2.0092 0.0000 1.4468 0.0000 0.0000
# * pheno_survival
mydata[[3]] |> head()
# time status
# TCGA-69-7978 4.40 0
# TCGA-62-8399 88.57 0
# TCGA-78-7539 25.99 0
# TCGA-73-4658 52.56 1
# TCGA-44-6775 23.16 0
# TCGA-44-2655 43.50 0We recommend using the zeallot package’s
%<-% function to assign values and rename them
simultaneously.
6.3 Setting up Python Environment
Some screening methods (e.g. Section 3.5 DEGAS) are built using Python and require an execution environment. Here is a function to help you set up a Python environment. Both Windows and Unix-like systems are supported.
# * This is an example of setting up a Python environment using conda
SetupPyEnv(
env_type = "conda",
env_name = "test-condaenv",
method = c("reticulate", "system","environment"), # choose one of them, default is "reticulate"
env_file = NULL # path to environment.yml file, used when method = "environment"
python_version = NULL,
packages = c(
"pandas" = "1.3",
"numpy" = "any"
),
recreate = FALSE, # whether to remove the existing environment and recreate it
use_conda_forge = TRUE,
verbose = TRUE
)
reticualte::use_condaenv("test-condaenv")
# * Or use virtualenv via reticulate
SetupPyEnv(
env_type = "venv",
env_name = "test-venv",
python_version = "3.9.15",
packages = c("tensorflow" = "2.4.1", "protobuf" = "3.20.3"),
python_path = NULL,
recreate = FALSE,
verbose = TRUE
)
reticulate::use_virtualenv("test-venv")You can use ListPyEnv() to list all the Python
environments you have set up. Both conda and virtual environments are
supported.
# * Unix-like systems
ListPyEnv()
# name python type
# 1 base /home/user/miniconda3/bin/python conda
# 2 test-condaenv /home/user/miniconda3/envs/test-condaenv/bin/python conda
# 3 test-venv /home/user/miniconda3/envs/test-venv/bin/python venvShow the conda environments only:
# * Unix-like systems
ListPyEnv(env_type = "conda")
# name python type
# 1 base /home/user/miniconda3/bin/python conda
# 2 test-condaenv /home/user/miniconda3/envs/test-condaenv/bin/python condaIf the virtual environment isn’t installed in the default location,
you can specify the location of the virtual environment with the
venv_locations parameter.
ListPyEnv(env_type = "venv",venv_locations ="~/here_is_a_dir/.virtualenvs")6.4 Finding the Optimal Number of Principle Components
Usually the number if principle components (PCs) is manually set to 10 or 20 according to the elbow plot. However, it is not always the case that the PCs with the highest variance are the most informative. Here is a function to help you find the optimal number of PCs.
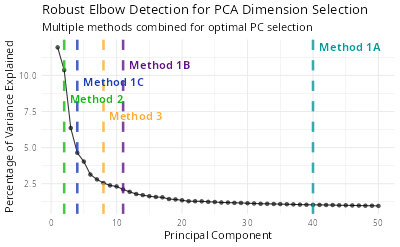
ndims <- FindRobustElbow(
obj = seurat,
verbose = TRUE,
ndims = 50
)
# ── Method Results
# ℹ Method 1A (Cumulative Variance > 90%): 1:40
# ℹ Method 1B (Variance > Mean): 1:11
# ℹ Method 1C (Variance > 2*SD): 1:4
# ℹ Method 2 (Second Derivative): 1:2
# ℹ Method 3 (Distance-based): 1:8
# ✔ Final Recommended Dimensions: 1:35
# ── Summary
# ℹ Cumulative variance at 35 PCs: 84.8%
# ℹ Variance explained by PC35: 1.09%
print(ndims)
# 35This function will draw an elbow plot with each method and the
recommended number of PCs. You can also use the ndims
parameter to specify the maximum number of PCs to be tested. The default
value is 50.
knitr::include_graphics("vignettes/example_figures/elbow.png")7. Troubleshooting
View Troubleshooting(https://github.com/WangX-Lab/SigBridgeR/wiki/Troubleshooting) for troubleshooting, especially for the environment setup.
Session information:
sessionInfo()
# R version 4.4.1 (2024-06-14)
# Platform: x86_64-pc-linux-gnu
# Running under: Ubuntu 24.04.3 LTS
# Matrix products: default
# BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
# LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=zh_CN.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=zh_CN.UTF-8
# [6] LC_MESSAGES=en_US.UTF-8 LC_PAPER=zh_CN.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=zh_CN.UTF-8 LC_IDENTIFICATION=C
# time zone: Asia/Shanghai
# tzcode source: system (glibc)
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
# other attached packages:
# [1] magrittr_2.0.4 dplyr_1.1.4
# loaded via a namespace (and not attached):
# [1] RColorBrewer_1.1-3 sys_3.4.3 jsonlite_2.0.0 spatstat.utils_3.2-0 farver_2.1.2 Scissor_2.0.0
# [7] vctrs_0.6.5 ROCR_1.0-11 spatstat.explore_3.5-3 memoise_2.0.1 RCurl_1.98-1.17 askpass_1.2.1
# [13] htmltools_0.5.8.1 BiocBaseUtils_1.6.0 curl_7.0.0 sctransform_0.4.2 parallelly_1.45.1 KernSmooth_2.23-24
# [19] htmlwidgets_1.6.4 ica_1.0-3 plyr_1.8.9 httr2_1.0.4 plotly_4.11.0 zoo_1.8-14
# [25] cachem_1.1.0 igraph_2.2.0 mime_0.13 lifecycle_1.0.4 pkgconfig_2.0.3 Matrix_1.7-4
# [31] R6_2.6.1 fastmap_1.2.0 BiocCheck_1.40.0 fitdistrplus_1.2-4 future_1.67.0 shiny_1.11.1
# [37] digest_0.6.37 patchwork_1.3.2 ps_1.8.0 tensor_1.5.1 Seurat_5.3.0 RSpectra_0.16-2
# [43] irlba_2.3.5.1 RSQLite_2.4.3 filelock_1.0.3 progressr_0.16.0 spatstat.sparse_3.1-0 polyclip_1.10-7
# [49] abind_1.4-8 httr_1.4.7 compiler_4.4.1 bit64_4.6.0-1 withr_3.0.2 S7_0.2.0
# [55] biocViews_1.72.0 DBI_1.2.3 chk_0.10.0 fastDummies_1.7.5 ggupset_0.4.1 ScPP_0.0.0.9000
# [61] MASS_7.3-60.2 openssl_2.3.4 rappdirs_0.3.3 tools_4.4.1 lmtest_0.9-40 httpuv_1.6.16
# [67] future.apply_1.20.0 goftest_1.2-3 glue_1.8.0 callr_3.7.6 nlme_3.1-164 promises_1.3.3
# [73] grid_4.4.1 stringdist_0.9.14 Rtsne_0.17 cluster_2.1.6 reshape2_1.4.4 generics_0.1.4
# [79] spatstat.data_3.1-8 gtable_0.3.6 preprocessCore_1.66.0 tidyr_1.3.1 data.table_1.17.8 sp_2.2-0
# [85] spatstat.geom_3.6-0 BiocGenerics_0.50.0 RcppAnnoy_0.0.22 stringr_1.5.2 ggrepel_0.9.6 RANN_2.6.2
# [91] pillar_1.11.1 spam_2.11-1 RcppHNSW_0.6.0 later_1.4.4 splines_4.4.1 scAB_1.0.0
# [97] BiocFileCache_2.12.0 lattice_0.22-6 deldir_2.0-4 survival_3.6-4 bit_4.6.0 tidyselect_1.2.1
# [103] RBGL_1.80.0 miniUI_0.1.2 pbapply_1.7-4 knitr_1.50 gridExtra_2.3 scattermore_1.2
# [109] stats4_4.4.1 xfun_0.53 Biobase_2.64.0 credentials_2.0.1 matrixStats_1.5.0 stringi_1.8.7
# [115] lazyeval_0.2.2 evaluate_1.0.5 codetools_0.2-20 tibble_3.3.0 BiocManager_1.30.26 graph_1.82.0
# [121] cli_3.6.5 uwot_0.2.3 xtable_1.8-4 reticulate_1.43.0 processx_3.8.4 dichromat_2.0-0.1
# [127] Rcpp_1.1.0 gert_2.1.2 spatstat.random_3.4-2 globals_0.18.0 dbplyr_2.5.1 png_0.1-8
# [133] spatstat.univar_3.1-4 XML_3.99-0.19 RUnit_0.4.33.1 parallel_4.4.1 ggplot2_4.0.0 blob_1.2.4
# [139] dotCall64_1.2 bitops_1.0-9 listenv_0.9.1 viridisLite_0.4.2 scales_1.4.0 ggridges_0.5.7
# [145] SeuratObject_5.2.0 purrr_1.1.0 rlang_1.1.6 cowplot_1.2.0 8. References
Sun D, Guan X, Moran AE, Wu LY, Qian DZ, Schedin P, et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat Biotechnol. 2022 Apr;40(4):527–38.
Xie A, Wang H, Zhao J, Wang Z, Xu J, Xu Y. scPAS: single-cell phenotype-associated subpopulation identifier. Briefings in Bioinformatics. 2024 Nov 22;26(1):bbae655.
Zhang Q, Jin S, Zou X. scAB detects multiresolution cell states with clinical significance by integrating single-cell genomics and bulk sequencing data. Nucleic Acids Research. 2022 Nov 28;50(21):12112–30.
WangX-Lab/ScPP Internet. cited 2025 Aug 31. Available from: https://github.com/WangX-Lab/ScPP
Johnson TS, Yu CY, Huang Z, Xu S, Wang T, Dong C, et al. Diagnostic Evidence GAuge of Single cells (DEGAS): a flexible deep transfer learning framework for prioritizing cells in relation to disease. Genome Med. 2022 Feb 1;14(1):11.