Quick Start Guide for SigBridgeR
if (!requireNamespace("remotes")) {
install.packages("remotes")
}
remotes::install_github("WangLabCSU/SigBridgeR")
# # Or install from r-universe:
# install.packages("SigBridgeR", repos = "https://wanglabcsu.r-universe.dev")
library(SigBridgeR)
library(Seurat)We will start with a simple example.
if (requireNamespace("here", quietly = TRUE)) {
setwd(here::here())
knitr::opts_knit$set(root.dir = here::here())
}
library(zeallot) # %<-%
c(mat_exam, bulk_bi, pheno_bi) %<-% LoadRefData(data_type = "binary")mat_exam is a single-cell RNA expression matrix,
bulk_bi is a bulk tissue RNA expression matrix, and
pheno_bi is the phenotypic data associated with
bulk_bi. When using a binary or continuous phenotype, the
reference phenotype data is a named vector.
head(pheno_bi)
# TCGA-CA-5256-01 TCGA-AZ-6599-01 TCGA-AA-3655-01 TCGA-A6-6137-01 TCGA-CK-4952-01 TCGA-A6-5657-01
# 1 1 1 1 1 1By the way, when usinng a survival phenotype, the reference data is a data.frame.
pheno_sur <- LoadRefData(data_type = "survival")[[3]]
head(pheno_sur)
# time status
# TCGA-69-7978 4.40 0
# TCGA-62-8399 88.57 0
# TCGA-78-7539 25.99 0
# TCGA-73-4658 52.56 1
# TCGA-44-6775 23.16 0
# TCGA-44-2655 43.50 0The single-cell RNA expression matrix needs to be processed into a Seurat object. We set scale_features to all genes in order to maximize the flexibility of downstream analyses and capture a broader range of biological signals, so as to avoid insignificant results caused by too small a dataset.
seurat_obj <- SCPreProcess(
mat_exam,
quality_control.pattern = "^MT-",
scale_features = rownames(mat_exam),
dims = 1:20
)Then we can use these data to screen out phenotype-assoicated cells. Let’s start by trying Scissor.
scissor_res <- Screen(
bulk_bi,
seurat_obj,
pheno_bi,
phenotype_class = "binary",
screen_method = "Scissor",
alpha = 0.05
)Other screening methods are also available.
scpas_res <- Screen(
bulk_bi,
seurat_obj,
pheno_bi,
phenotype_class = "binary",
screen_method = "scPAS",
alpha = 0.05
)Since the screening is performed on the same data, we merge them.
merged_seurat <- MergeResult(
scissor_res,
scpas_res
)Finally, we visualize the screening results.
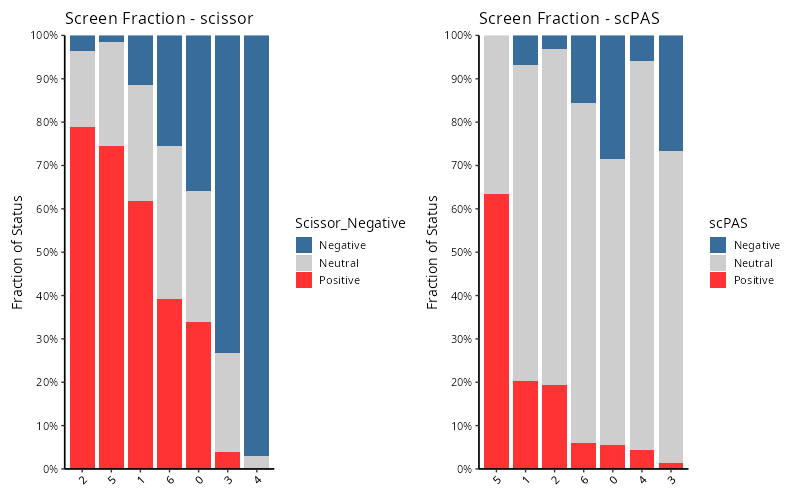
- stacked bar plot:
fraction = ScreenFractionPlot(
merged_seurat,
group_by = "seurat_clusters",
screen_type = c("scissor", "scPAS")
)
# names(fraction)
# [1] "stats" "plot" "combined_plot"
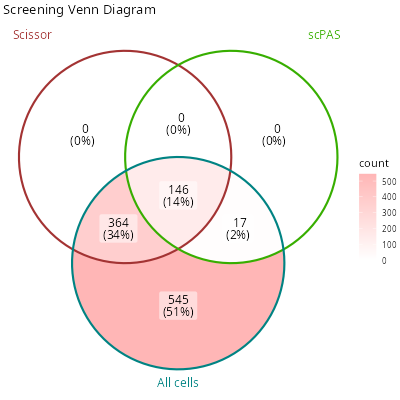
knitr::include_graphics("vignettes/example_figures/fraction_q.png")- Venn diagram:
c(scissor_pos, scpas_pos) %<-%
purrr::map(
c("scissor", "scPAS"),
~ colnames(merged_seurat)[
which(merged_seurat[[.x]] == "Positive")
]
)
all_cells <- colnames(seurat_obj)
pos_venn = list(
scissor = scissor_pos,
scpas = scpas_pos,
all_cells = all_cells
)
set.seed(123)
venn_plot = ggVennDiagram::ggVennDiagram(
x = pos_venn,
# * the labels of each group to be shown on the diagram
category.names = c(
"Scissor",
"scPAS",
"All cells"
),
# * the colors of each group
set_color = c(
"#a33333ff",
"#37ae00ff",
"#008383ff"
)
) +
ggplot2::scale_fill_gradient(low = "white", high = "#ffb6b6ff") +
ggplot2::ggtitle("Screening Venn Diagram")
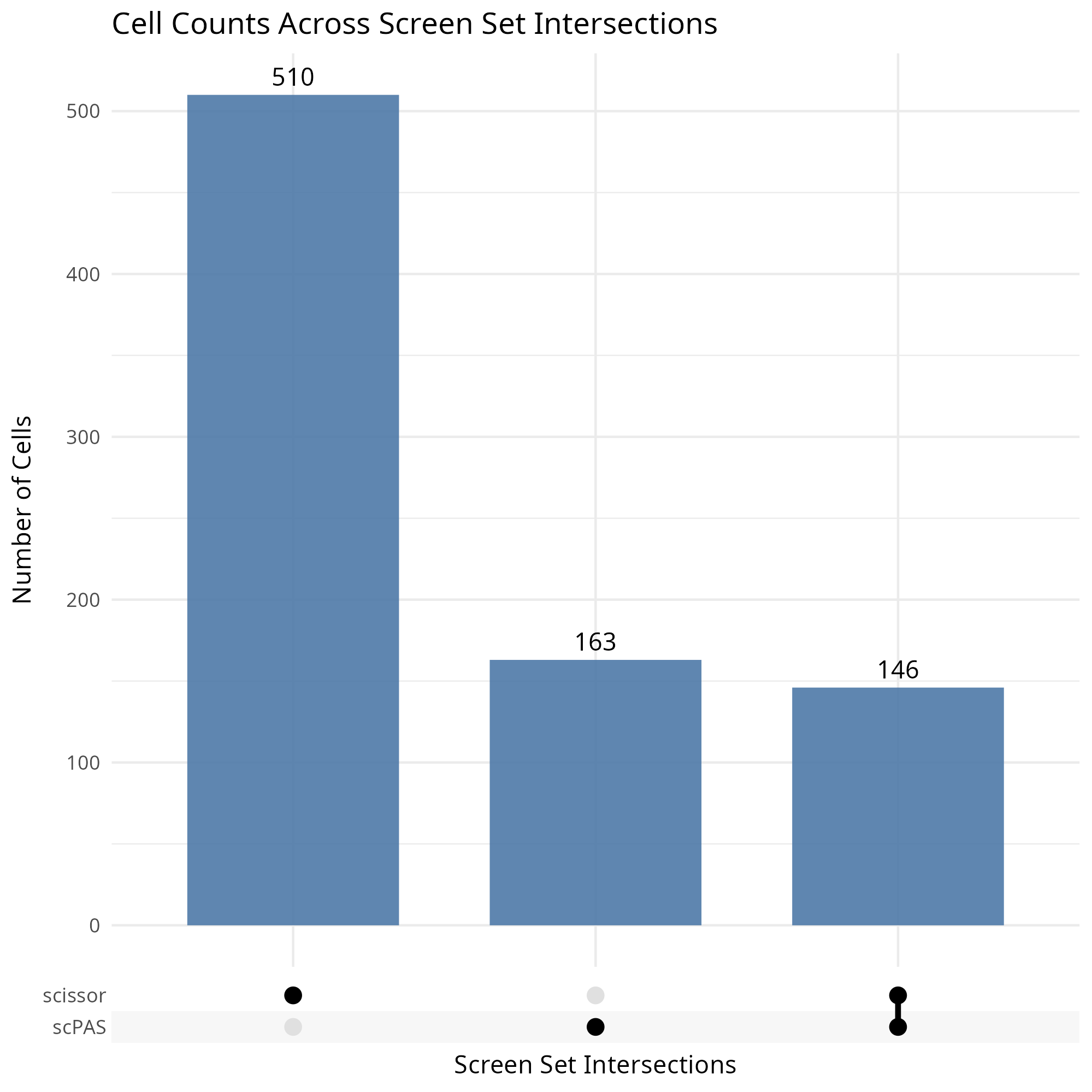
knitr::include_graphics("vignettes/example_figures/venn_q.png")- Set plot:
upset <- ScreenUpset(
merged_seurat,
screen_type = c("scissor", "scPAS")
)
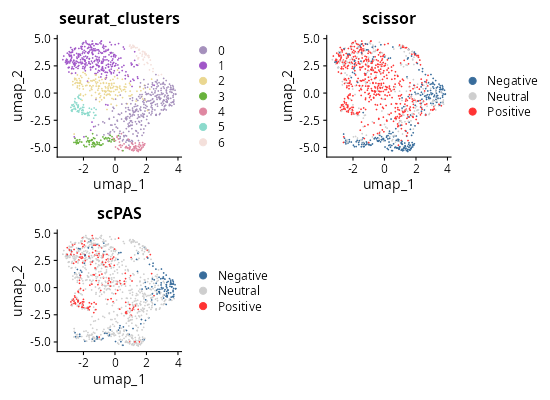
knitr::include_graphics("vignettes/example_figures/upset_q.png")- 2D UMAP:
library(patchwork)
library(randomcoloR)
c(
scissor_umap,
scpas_umap
) %<-%
purrr::map(
c("scissor", "scPAS"),
~ Seurat::DimPlot(
merged_seurat,
group.by = .x,
pt.size = 0.1,
reduction = "umap",
cols = c(
"Neutral" = "#CECECE",
"Positive" = "#ff3333",
"Negative" = "#386c9b"
)
) +
ggplot2::ggtitle(.x)
)
set.seed(123)
cols = randomcoloR::distinctColorPalette(
length(unique(merged_seurat$seurat_clusters)),
runTsne = TRUE
)
cluster_umap <- Seurat::DimPlot(
merged_seurat,
group.by = "seurat_clusters",
pt.size = 0.1,
reduction = "umap",
cols = cols
) +
ggplot2::ggtitle("seurat_clusters")
# * Show
umaps = cluster_umap +
scissor_umap +
scpas_umap +
plot_layout(ncol = 2)
umaps
knitr::include_graphics("vignettes/example_figures/umaps_q.png")Session information:
## R version 4.5.1 (2025-06-13)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## loaded via a namespace (and not attached):
## [1] digest_0.6.37 desc_1.4.3 R6_2.6.1 fastmap_1.2.0
## [5] xfun_0.53 cachem_1.1.0 knitr_1.50 htmltools_0.5.8.1
## [9] rmarkdown_2.30 lifecycle_1.0.4 cli_3.6.5 sass_0.4.10
## [13] pkgdown_2.1.3 textshaping_1.0.4 jquerylib_0.1.4 systemfonts_1.3.1
## [17] compiler_4.5.1 tools_4.5.1 ragg_1.5.0 bslib_0.9.0
## [21] evaluate_1.0.5 yaml_2.3.10 jsonlite_2.0.0 rlang_1.1.6
## [25] fs_1.6.6 htmlwidgets_1.6.4